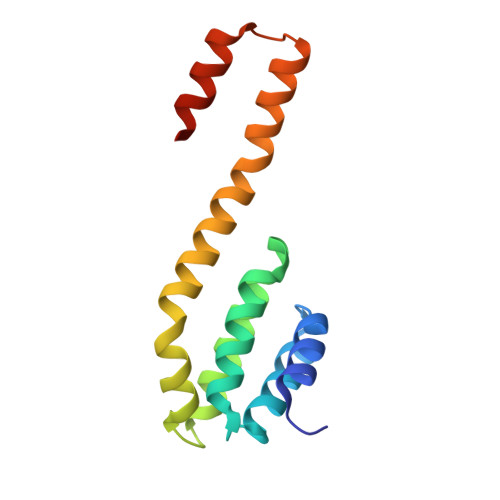
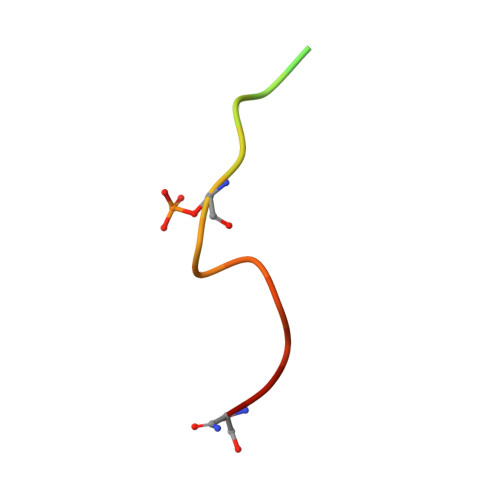
RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Ni, Z., Xu, C., Guo, X., Hunter, G.O., Kuznetsova, O.V., Tempel, W., Marcon, E., Zhong, G., Guo, H., Kuo, W.H., Li, J., Young, P., Olsen, J.B., Wan, C., Loppnau, P., El Bakkouri, M., Senisterra, G.A., He, H., Huang, H., Sidhu, S.S., Emili, A., Murphy, S., Mosley, A.L., Arrowsmith, C.H., Min, J., Greenblatt, J.F.(2014) Nat Struct Mol Biol 21: 686-695
- PubMed: 24997600
- DOI: https://doi.org/10.1038/nsmb.2853
- Primary Citation of Related Structures:
4FLA, 4FLB, 4JXT, 4Q94, 4Q96 - PubMed Abstract:
The RNA polymerase II (RNAPII) C-terminal domain (CTD) heptapeptide repeats (1-YSPTSPS-7) undergo dynamic phosphorylation and dephosphorylation during the transcription cycle to recruit factors that regulate transcription, RNA processing and chromatin modification. We show here that RPRD1A and RPRD1B form homodimers and heterodimers through their coiled-coil domains and interact preferentially via CTD-interaction domains (CIDs) with RNAPII CTD repeats phosphorylated at S2 and S7. Crystal structures of the RPRD1A, RPRD1B and RPRD2 CIDs, alone and in complex with RNAPII CTD phosphoisoforms, elucidate the molecular basis of CTD recognition. In an example of cross-talk between different CTD modifications, our data also indicate that RPRD1A and RPRD1B associate directly with RPAP2 phosphatase and, by interacting with CTD repeats where phospho-S2 and/or phospho-S7 bracket a phospho-S5 residue, serve as CTD scaffolds to coordinate the dephosphorylation of phospho-S5 by RPAP2.
- Donnelly Centre, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: