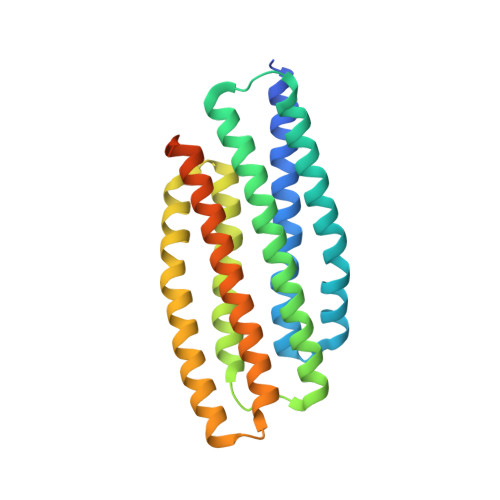
Crystal structure of PhoU from Pseudomonas aeruginosa, a negative regulator of the Pho regulon.
Lee, S.J., Park, Y.S., Kim, S.J., Lee, B.J., Suh, S.W.(2014) J Struct Biol 188: 22-29
- PubMed: 25220976
- DOI: https://doi.org/10.1016/j.jsb.2014.08.010
- Primary Citation of Related Structures:
4Q25 - PubMed Abstract:
In Escherichia coli, seven genes (pstS, pstC, pstA, pstB, phoU, phoR, and phoB) are involved in sensing environmental phosphate (Pi) and controlling the expression of the Pho regulon. PhoU is a negative regulator of the Pi-signaling pathway and modulates Pi transport through Pi transporter proteins (PstS, PstC, PstA, and PstB) through the two-component system PhoR and PhoB. Inactivation of PhoY2, one of the two PhoU homologs in Mycobacterium tuberculosis, causes defects in persistence phenotypes and increased susceptibility to antibiotics and stresses. Despite the important biological role, the mechanism of PhoU function is still unknown. Here we have determined the crystal structure of PhoU from Pseudomonas aeruginosa. It exists as a dimer in the crystal, with each monomer consisting of two structurally similar three-helix bundles. Our equilibrium sedimentation measurements support the reversible monomer-dimer equilibrium model in which P. aeruginosa PhoU exists in solution predominantly as dimers, with monomers in a minor fraction, at low protein concentrations. The dissociation constant for PhoU dimerization is 3.2×10(-6)M. The overall structure of P. aeruginosa PhoU dimer resembles those of Aquifex aeolicus PhoU and Thermotoga maritima PhoU2. However, it shows distinct structural features in some loops and the dimerization pattern.
- The Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul 151-742, Republic of Korea; Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 151-742, Republic of Korea.
Organizational Affiliation: