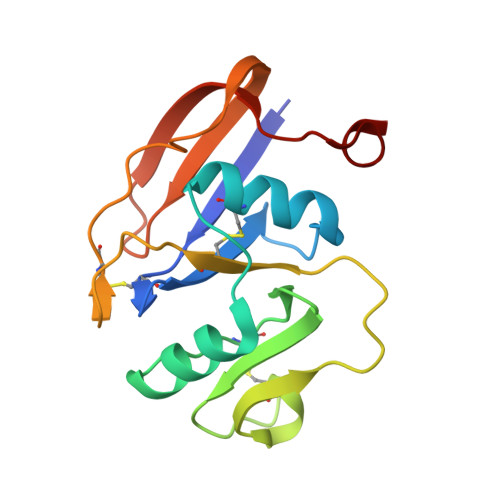
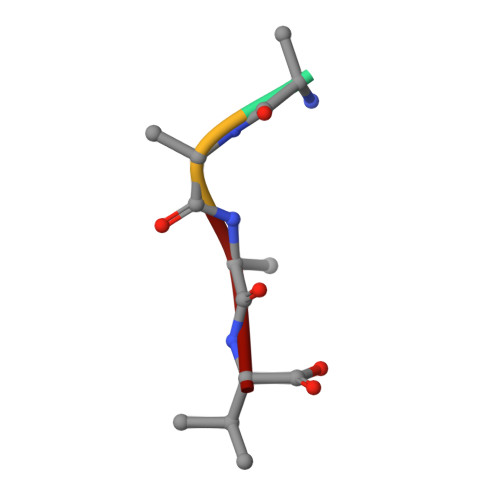
High-resolution crystal structures of alternate forms of the human CD44 hyaluronan-binding domain reveal a site for protein interaction.
Liu, L.K., Finzel, B.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 1155-1161
- PubMed: 25195884
- DOI: https://doi.org/10.1107/S2053230X14015532
- Primary Citation of Related Structures:
4PZ3, 4PZ4 - PubMed Abstract:
Two new crystal structures of the extracellular hyaluronan-binding domain of human CD44 are described at high resolution. A hexagonal crystal form at 1.60 Å resolution and a monoclinic form at 1.08 Å resolution both have two molecules in the asymmetric unit arranged about a similar noncrystallographic twofold axis of symmetry. These structures are compared with those previously reported at 2.20 Å resolution to show that the fold is quite resistant to structural deformation in different crystal environments. Unexpectedly, a short peptide is found in the monoclinic crystals at a site remote from the known hyaluronan-binding groove. The peptide with a valine at the carboxy-terminus must have co-purified from the bacterial expression host and binds on the opposite side of the domain from the known hyaluronan-binding groove. This opportunistic binding may identify a site of interaction used as CD44 assembles with other proteins to accomplish effective signaling regarding changes to the extracellular environment.
- Department of Medicinal Chemistry, University of Minnesota, 308 Harvard Street SE, Minneapolis, MN 55455, USA.
Organizational Affiliation: