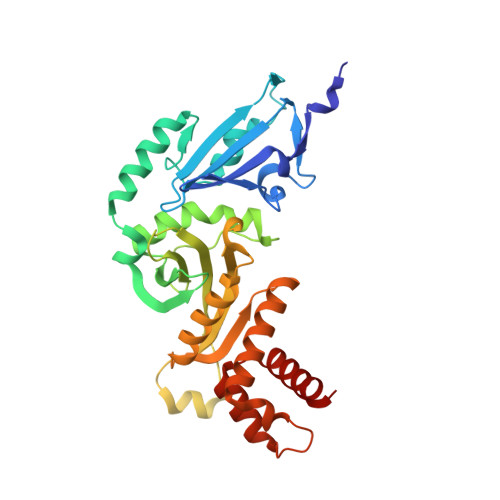
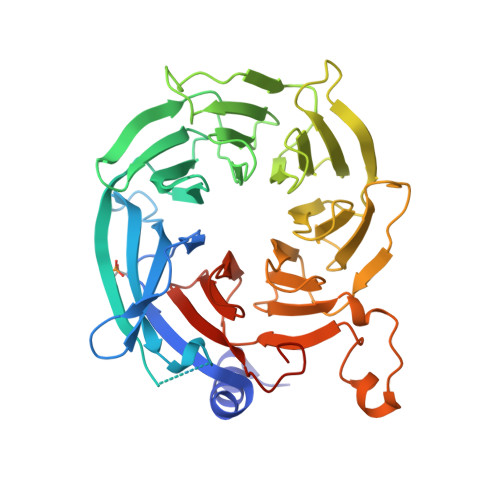
Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Li, Y., Zhang, L., Liu, T., Chai, C., Fang, Q., Wu, H., Agudelo garcia, P.A., Han, Z., Zong, S., Yu, Y., Zhang, X., Parthun, M.R., Chai, J., Xu, R.M., Yang, M.(2014) Genes Dev 28: 1217-1227
- PubMed: 24835250
- DOI: https://doi.org/10.1101/gad.240531.114
- Primary Citation of Related Structures:
4PSW, 4PSX - PubMed Abstract:
Post-translational modifications of histones are significant regulators of replication, transcription, and DNA repair. Particularly, newly synthesized histone H4 in H3/H4 heterodimers becomes acetylated on N-terminal lysine residues prior to its incorporation into chromatin. Previous studies have established that the histone acetyltransferase (HAT) complex Hat1p/Hat2p medicates this modification. However, the mechanism of how Hat1p/Hat2p recognizes and facilitates the enzymatic activities on the newly assembled H3/H4 heterodimer remains unknown. Furthermore, Hat2p is a WD40 repeat protein, which is found in many histone modifier complexes. However, how the WD40 repeat proteins facilitate enzymatic activities of histone modification enzymes is unclear. In this study, we first solved the high-resolution crystal structure of a Hat1p/Hat2p/CoA/H4 peptide complex and found that the H4 tail interacts with both Hat1p and Hat2p, by which substrate recruitment is facilitated. We further discovered that H3 N-terminal peptides can bind to the Hat2p WD40 domain and solved the structure of the Hat1p/Hat2p/CoA/H4/H3 peptide complex. Moreover, the interaction with Hat2p requires unmodified Arg2/Lys4 and Lys9 on the H3 tail, suggesting a novel model to specify the activity of Hat1p/Hat2p toward newly synthesized H3/H4 heterodimers. Together, our study demonstrated the substrate recognition mechanism by the Hat1p/Hat2p complex, which is critical for DNA replication and other chromatin remodeling processes.
- Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China;
Organizational Affiliation: