Coiled-coil Coactivators Play a Structural Role Mediating Interactions in Hypoxia-inducible Factor Heterodimerization.
Guo, Y., Scheuermann, T.H., Partch, C.L., Tomchick, D.R., Gardner, K.H.(2015) J Biological Chem 290: 7707-7721
- PubMed: 25627682
- DOI: https://doi.org/10.1074/jbc.M114.632786
- Primary Citation of Related Structures:
4LPZ, 4PKY - PubMed Abstract:
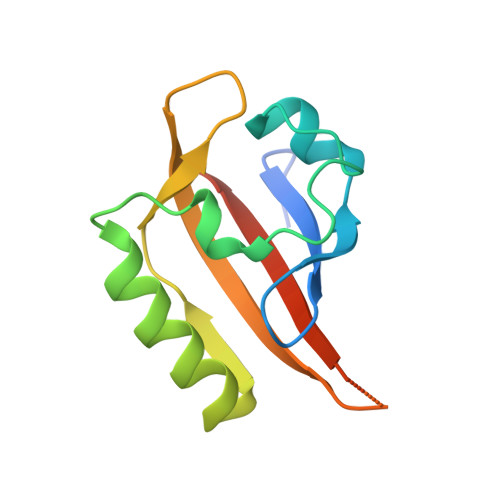
The hypoxia-inducible factor complex (HIF-α·aryl hydrocarbon receptor nuclear translocator (ARNT)) requires association with several transcription coactivators for a successful cellular response to hypoxic stress. In addition to the conventional global transcription coactivator CREB-binding protein/p300 (CBP/p300) that binds to the HIF-α transactivation domain, a new group of transcription coactivators called the coiled-coil coactivators (CCCs) interact directly with the second PER-ARNT-SIM (PAS) domain of ARNT (ARNT PAS-B). These less studied transcription coactivators play essential roles in the HIF-dependent hypoxia response, and CCC misregulation is associated with several forms of cancer. To better understand CCC protein recruitment by the heterodimeric HIF transcription factor, we used x-ray crystallography, NMR spectroscopy, and biochemical methods to investigate the structure of the ARNT PAS-B domain in complex with the C-terminal fragment of a coiled-coil coactivator protein, transforming acidic coiled-coil coactivator 3 (TACC3). We found that the HIF-2α PAS-B domain also directly interacts with TACC3, motivating an NMR data-derived model suggesting a means by which TACC3 could form a ternary complex with HIF-2α PAS-B and ARNT PAS-B via β-sheet/coiled-coil interactions. These findings suggest that TACC3 could be recruited as a bridge to cooperatively mediate between the HIF-2α PAS-B·ARNT PAS-B complex, thereby participating more directly in HIF-dependent gene transcription than previously anticipated.
- From the Departments of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, Dallas, Texas 75390-8816 and.
Organizational Affiliation: