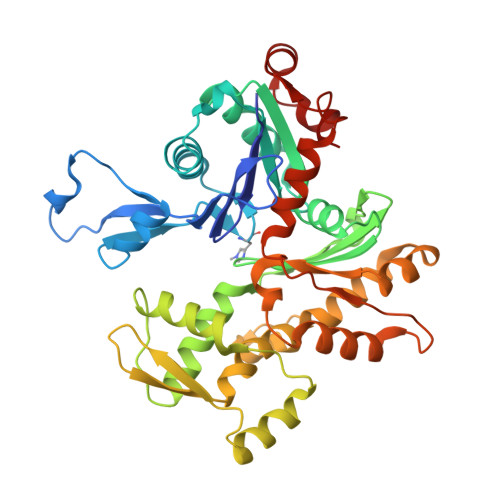
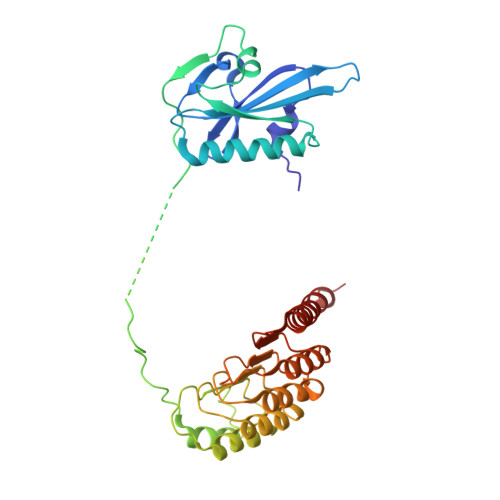
Actin cytoskeleton. Mechanism of actin filament pointed-end capping by tropomodulin.
Rao, J.N., Madasu, Y., Dominguez, R.(2014) Science 345: 463-467
- PubMed: 25061212
- DOI: https://doi.org/10.1126/science.1256159
- Primary Citation of Related Structures:
4PKG, 4PKH, 4PKI - PubMed Abstract:
Proteins that cap the ends of the actin filament are essential regulators of cytoskeleton dynamics. Whereas several proteins cap the rapidly growing barbed end, tropomodulin (Tmod) is the only protein known to cap the slowly growing pointed end. The lack of structural information severely limits our understanding of Tmod's capping mechanism. We describe crystal structures of actin complexes with the unstructured amino-terminal and the leucine-rich repeat carboxy-terminal domains of Tmod. The structures and biochemical analysis of structure-inspired mutants showed that one Tmod molecule interacts with three actin subunits at the pointed end, while also contacting two tropomyosin molecules on each side of the filament. We found that Tmod achieves high-affinity binding through several discrete low-affinity interactions, which suggests a mechanism for controlled subunit exchange at the pointed end.
- Department of Physiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Organizational Affiliation: