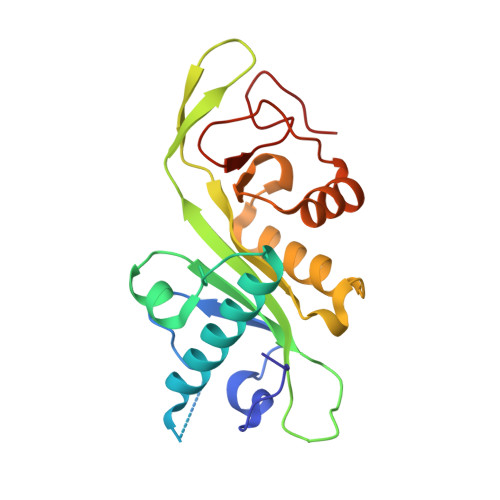
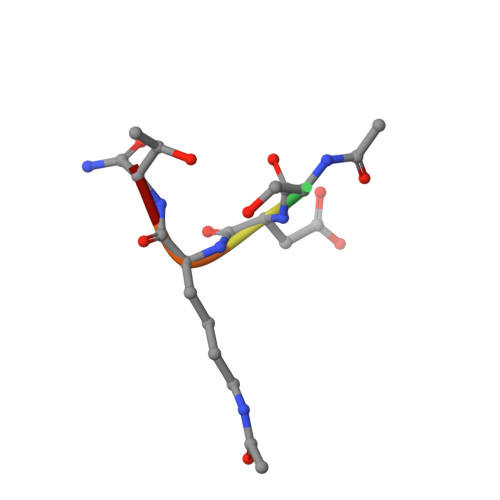
Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase.
Szyk, A., Deaconescu, A.M., Spector, J., Goodman, B., Valenstein, M.L., Ziolkowska, N.E., Kormendi, V., Grigorieff, N., Roll-Mecak, A.(2014) Cell 157: 1405-1415
- PubMed: 24906155
- DOI: https://doi.org/10.1016/j.cell.2014.03.061
- Primary Citation of Related Structures:
4PK2, 4PK3 - PubMed Abstract:
Acetylation of α-tubulin Lys40 by tubulin acetyltransferase (TAT) is the only known posttranslational modification in the microtubule lumen. It marks stable microtubules and is required for polarity establishment and directional migration. Here, we elucidate the mechanistic underpinnings for TAT activity and its preference for microtubules with slow turnover. 1.35 Å TAT cocrystal structures with bisubstrate analogs constrain TAT action to the microtubule lumen and reveal Lys40 engaged in a suboptimal active site. Assays with diverse tubulin polymers show that TAT is stimulated by microtubule interprotofilament contacts. Unexpectedly, despite the confined intraluminal location of Lys40, TAT efficiently scans the microtubule bidirectionally and acetylates stochastically without preference for ends. First-principles modeling and single-molecule measurements demonstrate that TAT catalytic activity, not constrained luminal diffusion, is rate limiting for acetylation. Thus, because of its preference for microtubules over free tubulin and its modest catalytic rate, TAT can function as a slow clock for microtubule lifetimes.
- Cell Biology and Biophysics Unit, Porter Neuroscience Research Center, National Institute of Neurological Disorders and Stroke, Bethesda, MD 20892, USA.
Organizational Affiliation: