Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1.
Jia, X., Weber, E., Tokarev, A., Lewinski, M., Rizk, M., Suarez, M., Guatelli, J., Xiong, Y.(2014) Elife 3: e02362-e02362
- PubMed: 24843023
- DOI: https://doi.org/10.7554/eLife.02362
- Primary Citation of Related Structures:
4P6Z - PubMed Abstract:
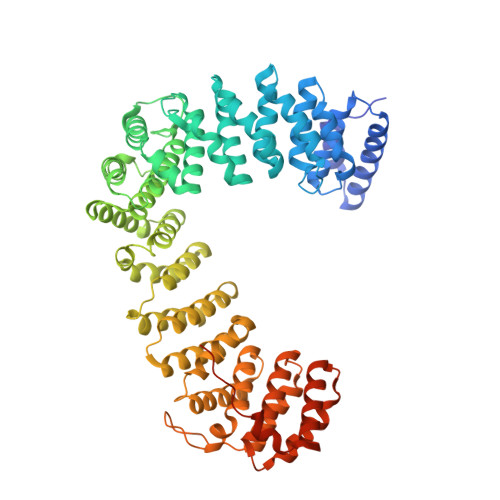
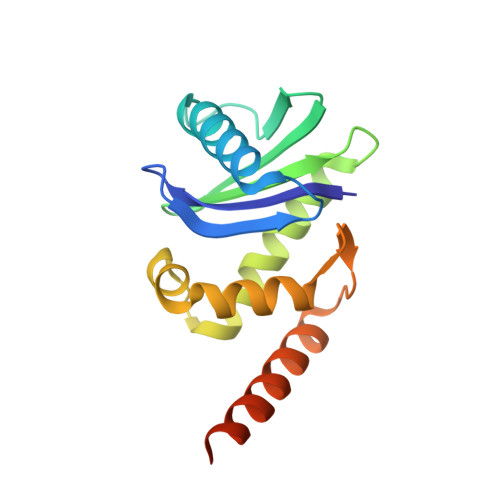
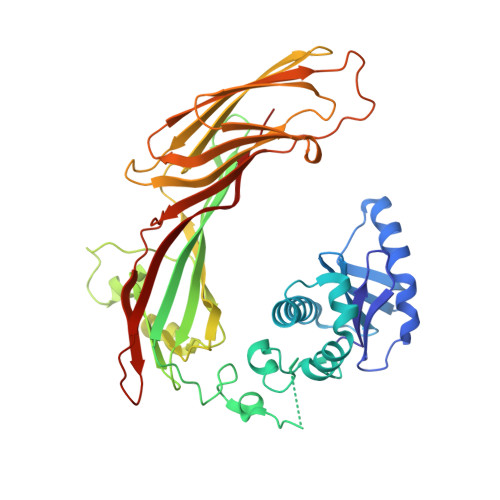
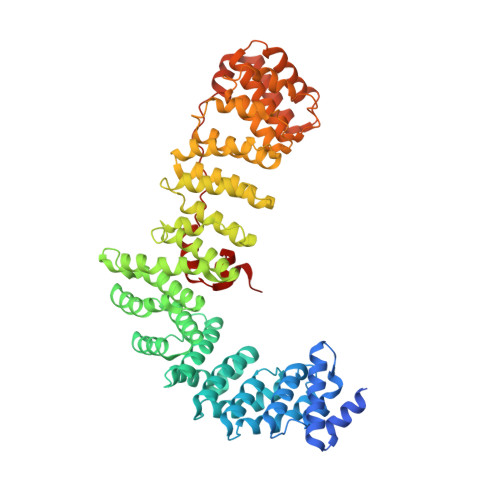
BST2/tetherin, an antiviral restriction factor, inhibits the release of enveloped viruses from the cell surface. Human immunodeficiency virus-1 (HIV-1) antagonizes BST2 through viral protein u (Vpu), which downregulates BST2 from the cell surface. We report the crystal structure of a protein complex containing Vpu and BST2 cytoplasmic domains and the core of the clathrin adaptor protein complex 1 (AP1). This, together with our biochemical and functional validations, reveals how Vpu hijacks the AP1-dependent membrane trafficking pathways to mistraffick BST2. Vpu mimics a canonical acidic dileucine-sorting motif to bind AP1 in the cytosol, while simultaneously interacting with BST2 in the membrane. These interactions enable Vpu to build on an intrinsic interaction between BST2 and AP1, presumably causing the observed retention of BST2 in juxtanuclear endosomes and stimulating its degradation in lysosomes. The ability of Vpu to hijack AP-dependent trafficking pathways suggests a potential common theme for Vpu-mediated downregulation of host proteins.DOI: http://dx.doi.org/10.7554/eLife.02362.001.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, United States.
Organizational Affiliation: