Crystal structure of the M42 aminopeptidase TmPep1050 from Thermotoga maritima
Dutoit, R., Brandt, N., Durisotti, V., Oudjama, Y., Demarez, M., Van Elder, D., Bauvois, C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aminopeptidase | 331 | Thermotoga maritima MSB8 | Mutation(s): 0 Gene Names: TM_1050, THEMA_09110, Tmari_1054 EC: 3.2.1.4 | 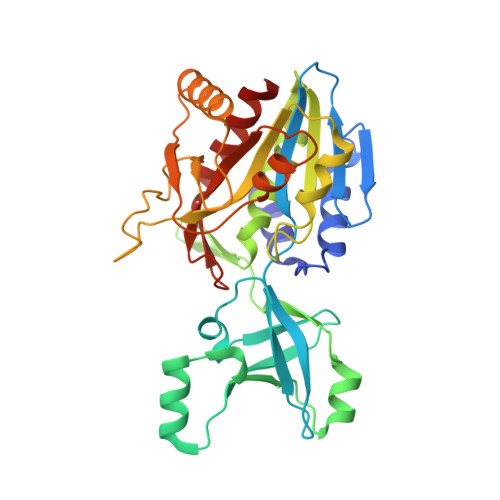 | |
UniProt | |||||
Find proteins for Q9X0E0 (Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)) Explore Q9X0E0 Go to UniProtKB: Q9X0E0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9X0E0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 114.26 | α = 114.46 |
| b = 114.57 | β = 91.71 |
| c = 114.04 | γ = 105.69 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Fonds de la Recherche Scientifique (FNRS) | Belgium | IISN 4.4505.00 |