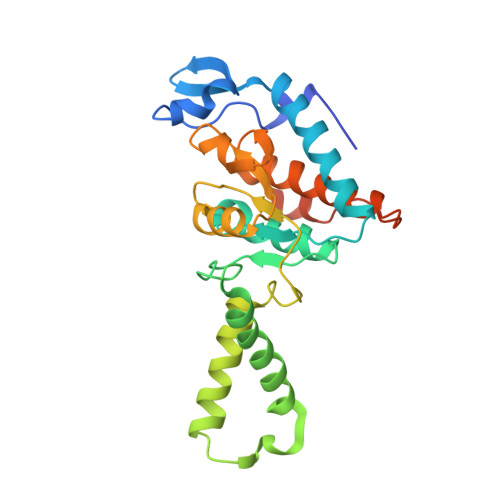
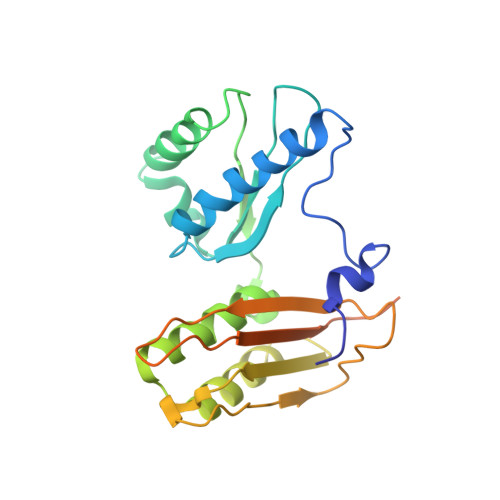
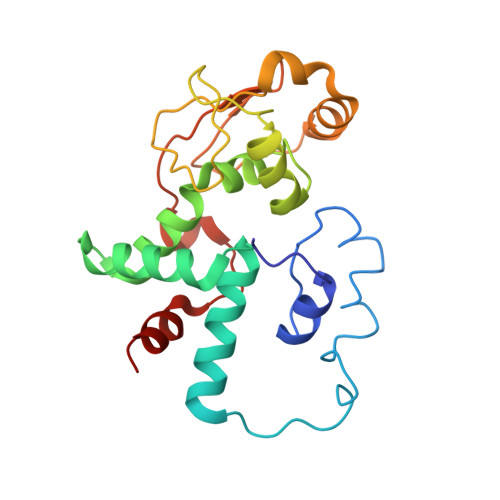
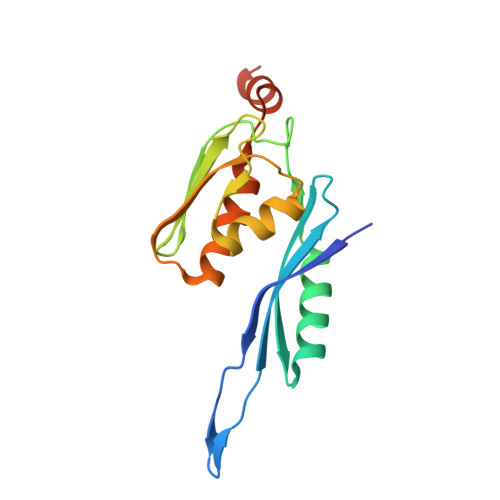
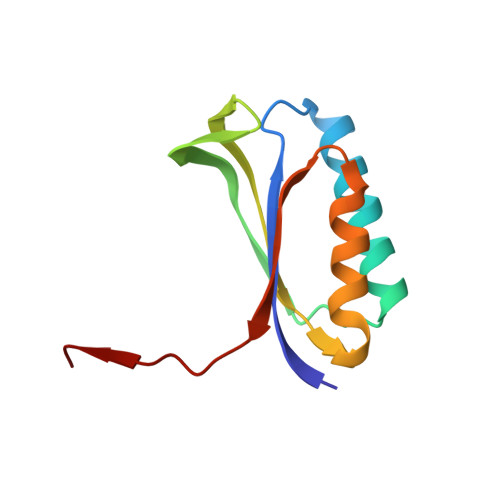
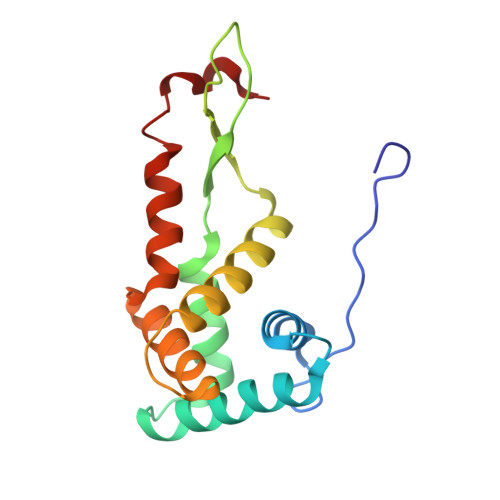
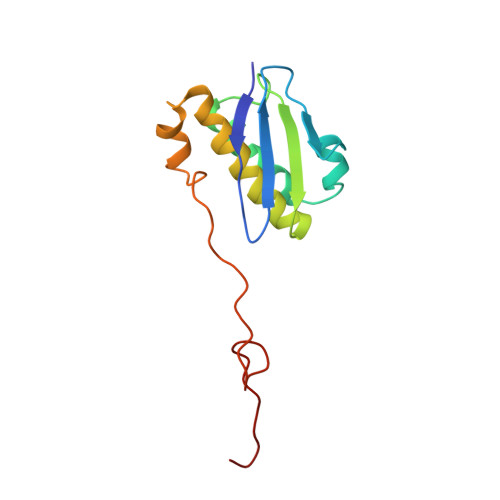
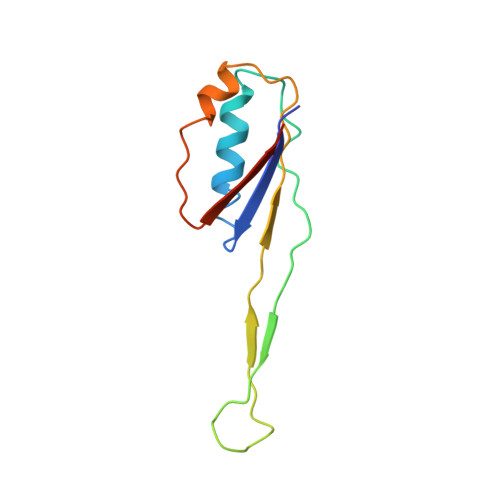
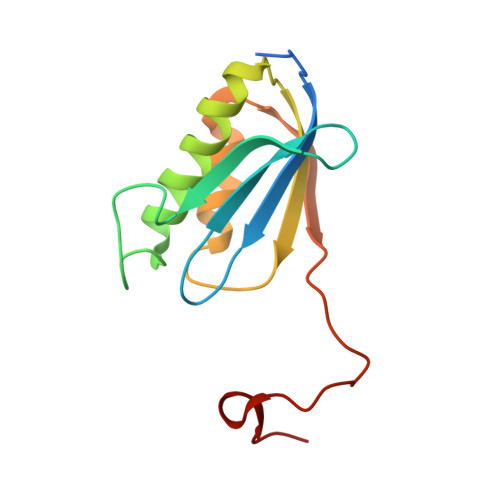
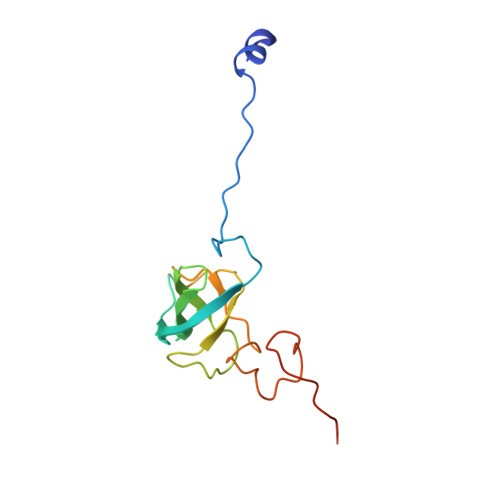
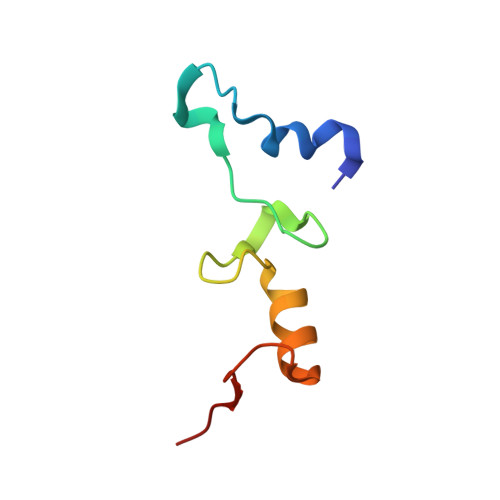
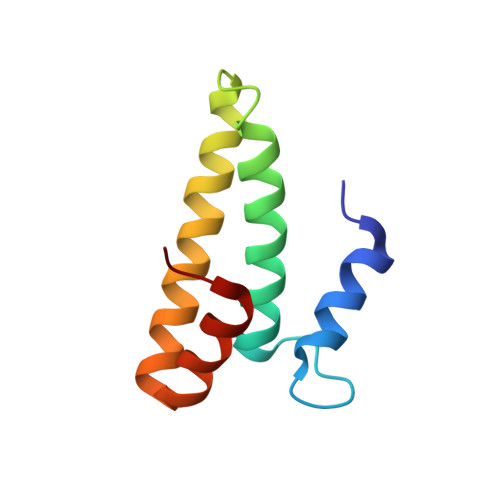
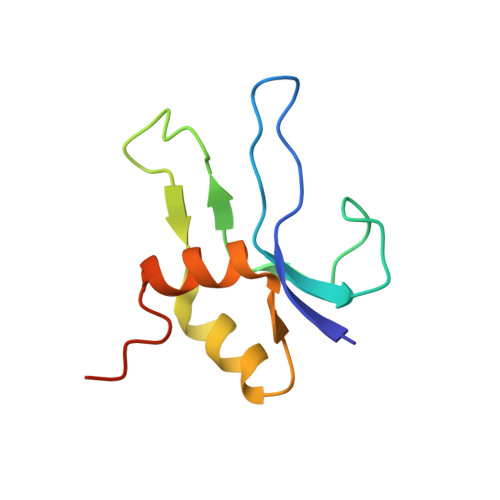
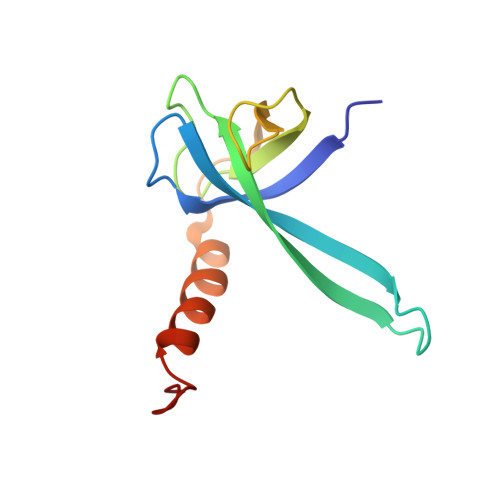
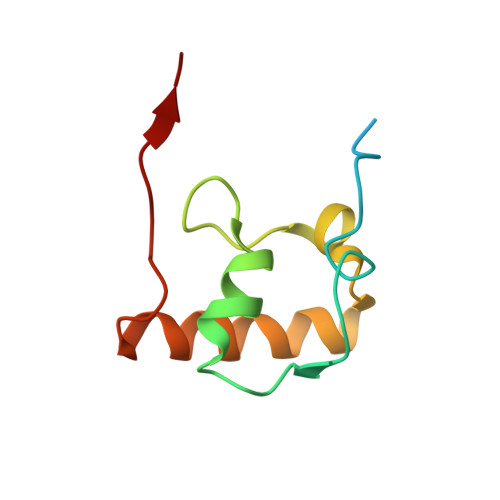
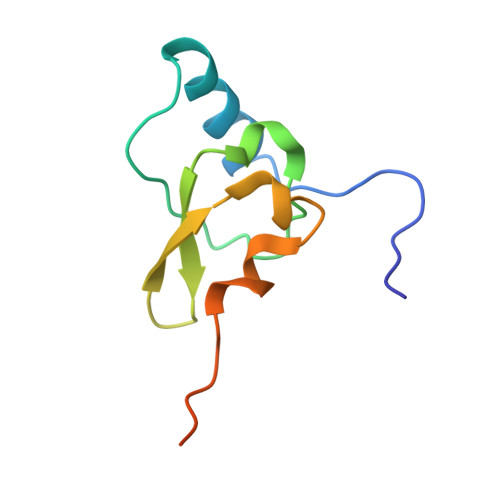
Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Dunkle, J.A., Vinal, K., Desai, P.M., Zelinskaya, N., Savic, M., West, D.M., Conn, G.L., Dunham, C.M.(2014) Proc Natl Acad Sci U S A 111: 6275-6280
- PubMed: 24717845
- DOI: https://doi.org/10.1073/pnas.1402789111
- Primary Citation of Related Structures:
4OX9 - PubMed Abstract:
Aminoglycosides are potent, broad spectrum, ribosome-targeting antibacterials whose clinical efficacy is seriously threatened by multiple resistance mechanisms. Here, we report the structural basis for 30S recognition by the novel plasmid-mediated aminoglycoside-resistance rRNA methyltransferase A (NpmA). These studies are supported by biochemical and functional assays that define the molecular features necessary for NpmA to catalyze m(1)A1408 modification and confer resistance. The requirement for the mature 30S as a substrate for NpmA is clearly explained by its recognition of four disparate 16S rRNA helices brought into proximity by 30S assembly. Our structure captures a "precatalytic state" in which multiple structural reorganizations orient functionally critical residues to flip A1408 from helix 44 and position it precisely in a remodeled active site for methylation. Our findings provide a new molecular framework for the activity of aminoglycoside-resistance rRNA methyltransferases that may serve as a functional paradigm for other modification enzymes acting late in 30S biogenesis.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322.
Organizational Affiliation: