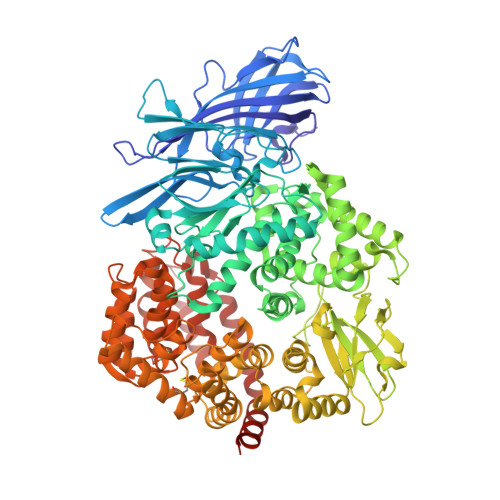
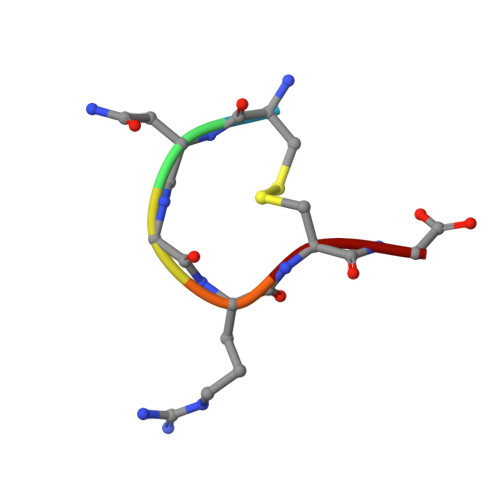
A Unified Mechanism for Aminopeptidase N-based Tumor Cell Motility and Tumor-homing Therapy.
Liu, C., Yang, Y., Chen, L., Lin, Y.L., Li, F.(2014) J Biological Chem 289: 34520-34529
- PubMed: 25359769
- DOI: https://doi.org/10.1074/jbc.M114.566802
- Primary Citation of Related Structures:
4OU3 - PubMed Abstract:
Tumor cell surface aminopeptidase N (APN or CD13) has two puzzling functions unrelated to its enzymatic activity: mediating tumor cell motility and serving as a receptor for tumor-homing peptides (peptides that bring anti-cancer drugs to tumor cells). To investigate APN-based tumor-homing therapy, we determined the crystal structure of APN complexed with a tumor-homing peptide containing a representative Asn-Gly-Arg (NGR) motif. The tumor-homing peptide binds to the APN enzymatic active site, but it resists APN degradation due to a distorted scissile peptide bond. To explore APN-based tumor cell motility, we examined the interactions between APN and extracellular matrix (ECM) proteins. APN binds to, but does not degrade, NGR motifs in ECM proteins that share similar conformations with the NGR motif in the APN-bound tumor-homing peptide. Therefore, APN-based tumor cell motility and tumor-homing therapy rely on a unified mechanism in which both functions are driven by the specific and stable interactions between APN and the NGR motifs in ECM proteins and tumor-homing peptides. This study further implicates APN as an integrin-like molecule that functions broadly in cell motility and adhesion by interacting with its signature NGR motifs in the extracellular environment.
- From the Department of Pharmacology, University of Minnesota Medical School, Minneapolis, Minnesota 55455.
Organizational Affiliation: