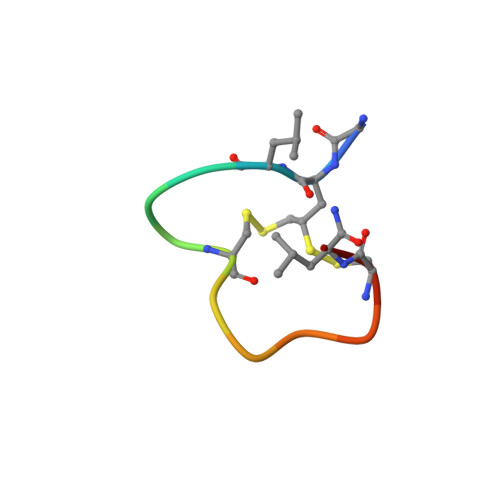
Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Chen, S., Gopalakrishnan, R., Schaer, T., Marger, F., Hovius, R., Bertrand, D., Pojer, F., Heinis, C.(2014) Nat Chem 6: 1009-1016
- PubMed: 25343607
- DOI: https://doi.org/10.1038/nchem.2043
- Primary Citation of Related Structures:
2MOA, 4OS1, 4OS2, 4OS4, 4OS5, 4OS6, 4OS7 - PubMed Abstract:
The disulfide bonds that form between two cysteine residues are important in defining and rigidifying the structures of proteins and peptides. In polypeptides containing multiple cysteine residues, disulfide isomerization can lead to multiple products with different biological activities. Here, we describe the development of a dithiol amino acid (Dtaa) that can form two disulfide bridges at a single amino acid site. Application of Dtaas to a serine protease inhibitor and a nicotinic acetylcholine receptor inhibitor that contain disulfide constraints enhanced their inhibitory activities 40- and 7.6-fold, respectively. X-ray crystallographic and NMR structure analysis show that the peptide ligands containing Dtaas have retained their native tertiary structures. We furthermore show that replacement of two cysteines by Dtaas can avoid the formation of disulfide bond isomers. With these properties, Dtaas are likely to have broad application in the rational design or directed evolution of peptides and proteins with high activity and stability.
- Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland.
Organizational Affiliation: