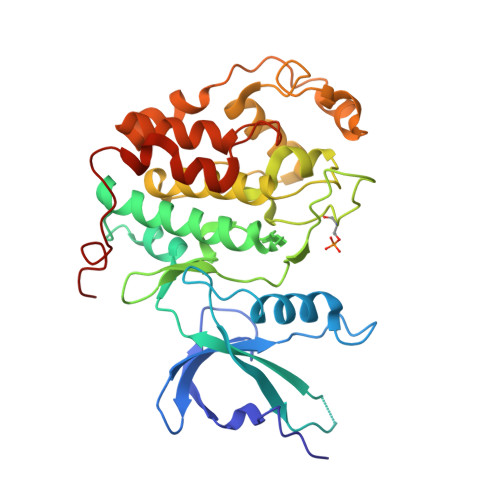
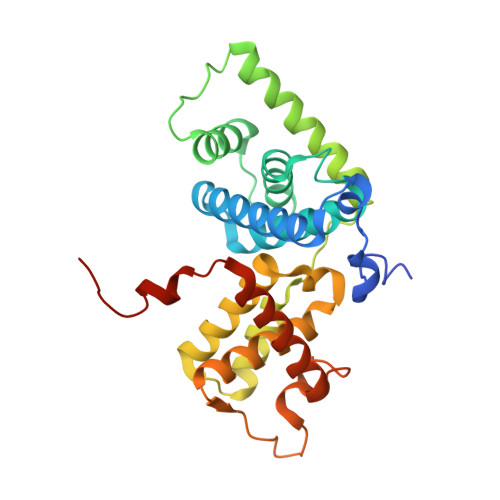
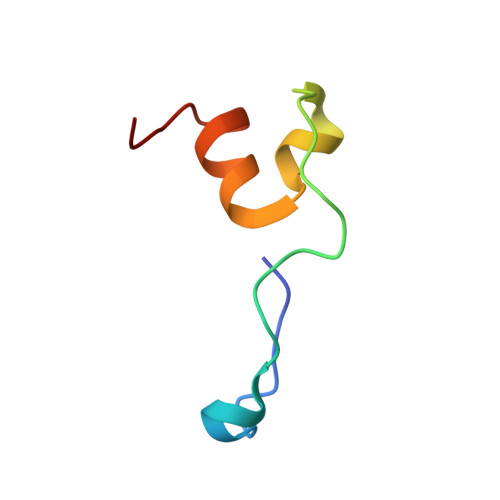
Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Gu, J., Babayeva, N.D., Suwa, Y., Baranovskiy, A.G., Price, D.H., Tahirov, T.H.(2014) Cell Cycle 13: 1788-1797
- PubMed: 24727379
- DOI: https://doi.org/10.4161/cc.28756
- Primary Citation of Related Structures:
4OR5 - PubMed Abstract:
Developing anti-viral therapies targeting HIV-1 transcription has been hampered by the limited structural knowledge of the proteins involved. HIV-1 hijacks the cellular machinery that controls RNA polymerase II elongation through an interaction of HIV-1 Tat with the positive transcription elongation factor P-TEFb, which interacts with an AF4 family member (AFF1/2/3/4) in the super elongation complex (SEC). Because inclusion of Tat•P-TEFb into the SEC is critical for HIV transcription, we have determined the crystal structure of the Tat•AFF4•P-TEFb complex containing HIV-1 Tat (residues 1-48), human Cyclin T1 (1-266), human Cdk9 (7-332), and human AFF4 (27-69). Tat binding to AFF4•P-TEFb causes concerted structural changes in AFF4 via a shift of helix H5' of Cyclin T1 and the α-3 10 helix of AFF4. The interaction between Tat and AFF4 provides structural constraints that explain tolerated Tat mutations. Analysis of the Tat-binding surface of AFF4 coupled with modeling of all other AF4 family members suggests that AFF1 and AFF4 would be preferred over AFF2 or AFF3 for interaction with Tat•P-TEFb. The structure establishes that the Tat-TAR recognition motif (TRM) in Cyclin T1 interacts with both Tat and AFF4, leading to the exposure of arginine side chains for binding to TAR RNA. Furthermore, modeling of Tat Lys28 acetylation suggests that the acetyl group would be in a favorable position for H-bond formation with Asn257 of TRM, thereby stabilizing the TRM in Cyclin T1, and provides a structural basis for the modulation of TAR RNA binding by acetylation of Tat Lys28.
- Eppley Institute for Research in Cancer and Allied Diseases; University of Nebraska Medical Center; Omaha, NE USA.
Organizational Affiliation: