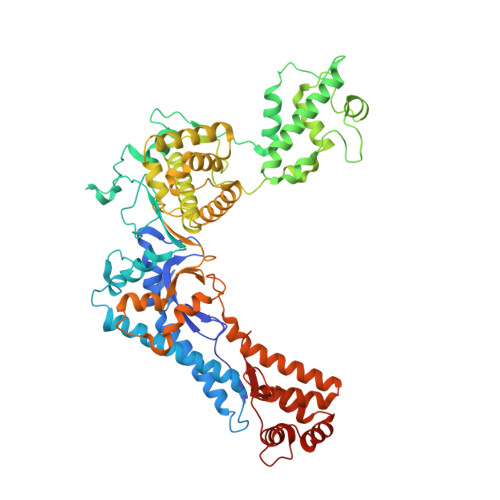
The structure of the N-terminal domain of the Legionella protein SidC
Gazdag, E.M., Schobel, S., Shkumatov, A.V., Goody, R.S., Itzen, A.(2014) J Struct Biol 186: 188-194
- PubMed: 24556577
- DOI: https://doi.org/10.1016/j.jsb.2014.02.003
- Primary Citation of Related Structures:
4OOJ - PubMed Abstract:
The Gram-negative bacterium Legionella pneumophila is the causative agent of Legionnaires' disease. During infection of eukaryotic cells, the bacterium releases about 300 different bacterial effector molecules that aid in the establishment of the Legionella-containing vacuole (LCV) among which SidC is one of these secreted proteins. However, apart from membrane lipid binding the function of SidC remains elusive. In order to characterize SidC further, we have determined the crystal structure of the N-terminal domain of SidC (amino acids 1-609, referred to as SidC-N) at 2.4Å resolution. SidC-N reveals a novel fold in which 4 potential subdomains (A-D) are arranged in a crescent-like structure. None of these subdomains currently has any known structural homologues, raising the question of how this fold has evolved. These domains are highly interconnected, with a low degree of flexibility towards each other. Due to the extended arrangement of the subdomains, SidC-N may contain multiple binding sites for potential interaction partners.
- Max Planck Institute of Molecular Physiology, Department of Physical Biochemistry, Dortmund 44227, Germany.
Organizational Affiliation: