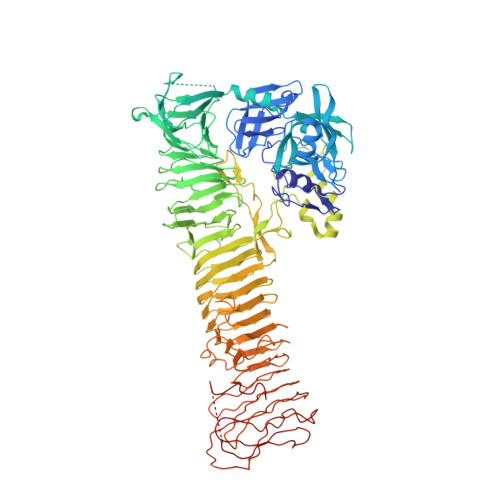
X-ray crystal structure of the passenger domain of plasmid encoded toxin(Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC).
Domingo Meza-Aguilar, J., Fromme, P., Torres-Larios, A., Mendoza-Hernandez, G., Hernandez-Chinas, U., Arreguin-Espinosa de Los Monteros, R.A., Eslava Campos, C.A., Fromme, R.(2014) Biochem Biophys Res Commun 445: 439-444
- PubMed: 24530907
- DOI: https://doi.org/10.1016/j.bbrc.2014.02.016
- Primary Citation of Related Structures:
4OM9 - PubMed Abstract:
Autotransporters (ATs) represent a superfamily of proteins produced by a variety of pathogenic bacteria, which include the pathogenic groups of Escherichia coli (E. coli) associated with gastrointestinal and urinary tract infections. We present the first X-ray structure of the passenger domain from the Plasmid-encoded toxin (Pet) a 100 kDa protein at 2.3 Å resolution which is a cause of acute diarrhea in both developing and industrialized countries. Pet is a cytoskeleton-altering toxin that induces loss of actin stress fibers. While Pet (pdb code: 4OM9) shows only a sequence identity of 50% compared to the closest related protein sequence, extracellular serine protease plasmid (EspP) the structural features of both proteins are conserved. A closer structural look reveals that Pet contains a β-pleaded sheet at the sequence region of residues 181-190, the corresponding structural domain in EspP consists of a coiled loop. Secondary, the Pet passenger domain features a more pronounced beta sheet between residues 135 and 143 compared to the structure of EspP.
- Departamento de Salud Pública Facultad de Medicina UNAM, Ciudad Universitaria Coyoacán 04510, D.F., Mexico; Laboratorio de Patogenicidad Bacteriana, Unidad de Hemato Oncología e Investigación, Hospital Infantil de México Federico Gómez 06720, D.F., Mexico.
Organizational Affiliation: