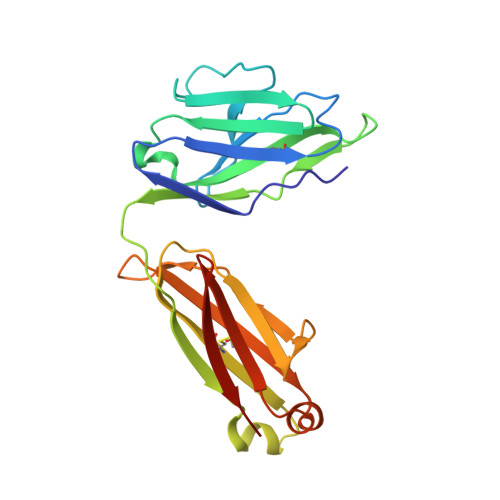
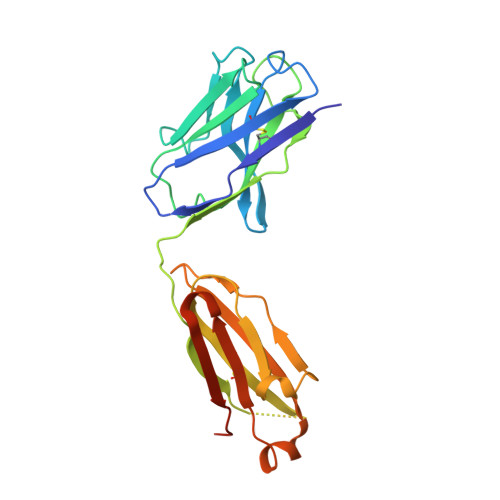
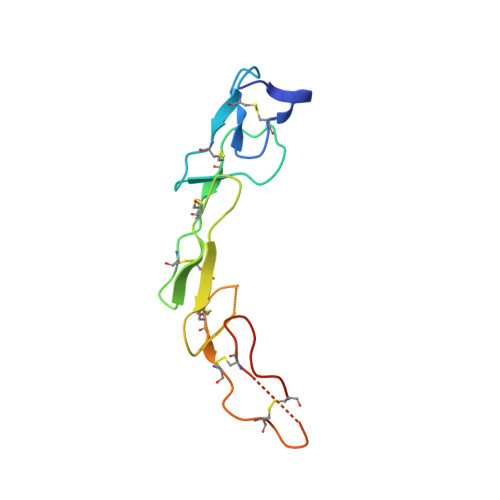
Structural and functional analysis of the interaction between the agonistic monoclonal antibody Apomab and the proapoptotic receptor DR5.
Adams, C., Totpal, K., Lawrence, D., Marsters, S., Pitti, R., Yee, S., Ross, S., Deforge, L., Koeppen, H., Sagolla, M., Compaan, D., Lowman, H., Hymowitz, S., Ashkenazi, A.(2008) Cell Death Differ 15: 751-761
- PubMed: 18219321
- DOI: https://doi.org/10.1038/sj.cdd.4402306
- Primary Citation of Related Structures:
4OD2 - PubMed Abstract:
Activation of the proapoptotic receptor death receptor5 (DR5) in various cancer cells triggers programmed cell death through the extrinsic pathway. We have generated a fully human monoclonal antibody (Apomab) that induces tumor cell apoptosis through DR5 and investigated the structural features of its interaction with DR5. Biochemical studies showed that Apomab binds DR5 tightly and selectively. X-ray crystallographic analysis of the complex between the Apomab Fab fragment and the DR5 ectodomain revealed an interaction epitope that partially overlaps with both regions of the Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand binding site. Apomab induced DR5 clustering at the cell surface and stimulated a death-inducing signaling complex containing the adaptor molecule Fas-associated death domain and the apoptosis-initiating protease caspase-8. Fc crosslinking further augmented Apomab's proapoptotic activity. In vitro, Apomab triggered apoptosis in cancer cells, while sparing normal hepatocytes even upon anti-Fc crosslinking. In vivo, Apomab exerted potent antitumor activity as a single agent or in combination with chemotherapy in xenograft models, including those based on colorectal, non-small cell lung and pancreatic cancer cell lines. These results provide structural and functional insight into the interaction of Apomab with DR5 and support further investigation of this antibody for cancer therapy.
- Department of Antibody Engineering, Genentech Inc., South San Francisco, CA 94080-4918, USA.
Organizational Affiliation: