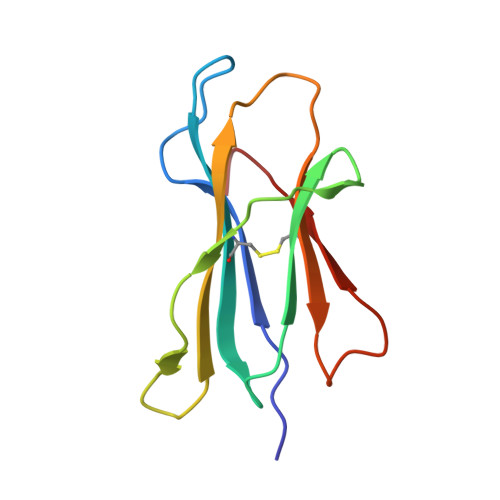
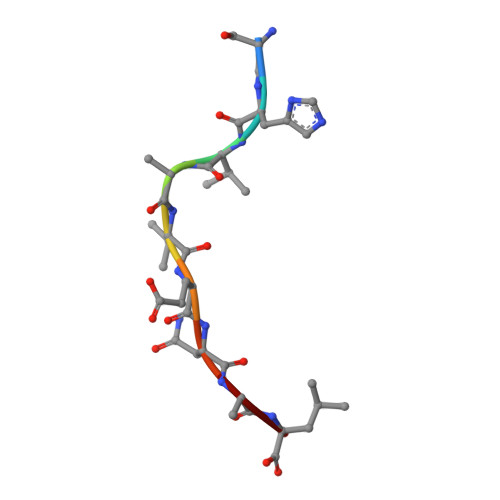
N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
Sun, M., Liu, J., Qi, J., Tefsen, B., Shi, Y., Yan, J., Gao, G.F.(2014) J Immunol 192: 5509-5519
- PubMed: 24829406
- DOI: https://doi.org/10.4049/jimmunol.1400199
- Primary Citation of Related Structures:
4O2C, 4O2E, 4O2F - PubMed Abstract:
As one of the most common posttranslational modifications (PTMs) of eukaryotic proteins, N(α)-terminal acetylation (Nt-acetylation) generates a class of N(α)-acetylpeptides that are known to be presented by MHC class I at the cell surface. Although such PTM plays a pivotal role in adjusting proteolysis, the molecular basis for the presentation and T cell recognition of N(α)-acetylpeptides remains largely unknown. In this study, we determined a high-resolution crystallographic structure of HLA (HLA)-B*3901 complexed with an N(α)-acetylpeptide derived from natural cellular processing, also in comparison with the unmodified-peptide complex. Unlike the α-amino-free P1 residues of unmodified peptide, of which the α-amino group inserts into pocket A of the Ag-binding groove, the N(α)-linked acetyl of the acetylated P1-Ser protrudes out of the groove for T cell recognition. Moreover, the Nt-acetylation not only alters the conformation of the peptide but also switches the residues in the α1-helix of HLA-B*3901, which may impact the T cell engagement. The thermostability measurements of complexes between N(α)-acetylpeptides and a series of MHC class I molecules derived from different species reveal reduced stability. Our findings provide the insight into the mode of N(α)-acetylpeptide-specific presentation by classical MHC class I molecules and shed light on the potential of acetylepitope-based immune intervene and vaccine development.
- School of Life Sciences, University of Science and Technology of China, Hefei 230027, China; CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China;
Organizational Affiliation: