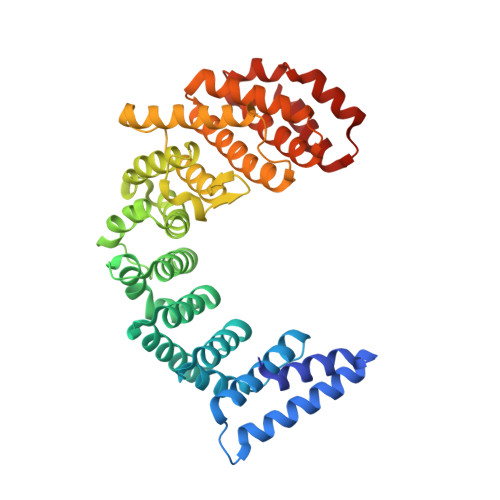How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4.
Ren, X., Park, S.Y., Bonifacino, J.S., Hurley, J.H.(2014) Elife 3: e01754-e01754
- PubMed: 24473078
- DOI: https://doi.org/10.7554/eLife.01754
- Primary Citation of Related Structures:
4NEE - PubMed Abstract:
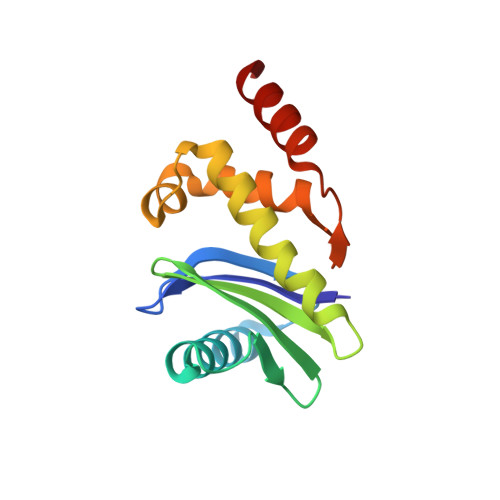
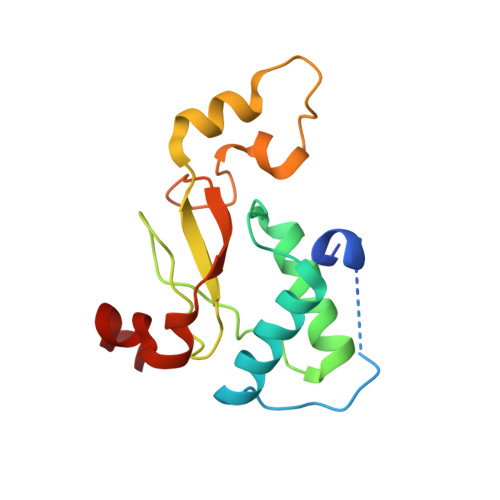
The Nef protein of HIV-1 downregulates the cell surface co-receptor CD4 by hijacking the clathrin adaptor complex AP-2. The structural basis for the hijacking of AP-2 by Nef is revealed by a 2.9 Å crystal structure of Nef bound to the α and σ2 subunits of AP-2. Nef binds to AP-2 via its central loop (residues 149-179) and its core. The determinants for Nef binding include residues that directly contact AP-2 and others that stabilize the binding-competent conformation of the central loop. Residues involved in both direct and indirect interactions are required for the binding of Nef to AP-2 and for downregulation of CD4. These results lead to a model for the docking of the full AP-2 tetramer to membranes as bound to Nef, such that the cytosolic tail of CD4 is situated to interact with its binding site on Nef. DOI: http://dx.doi.org/10.7554/eLife.01754.001.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States.
Organizational Affiliation: