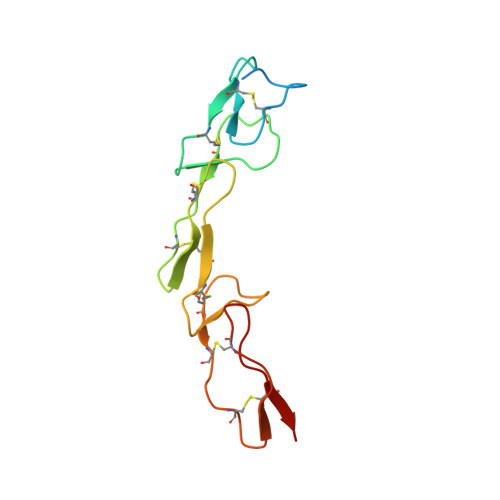
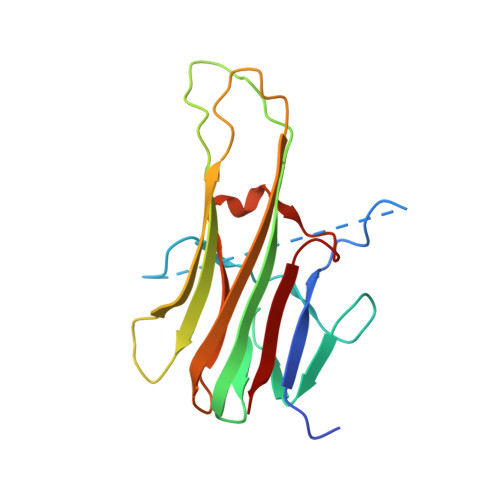
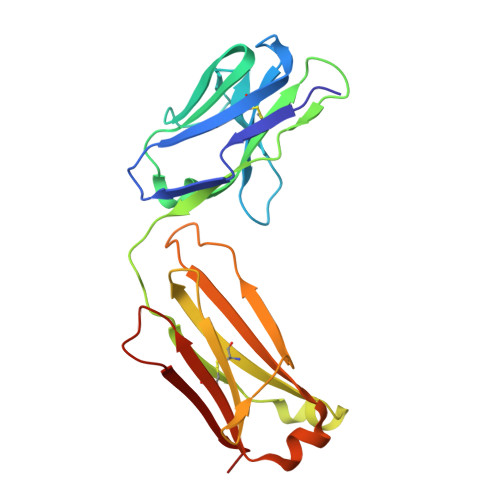
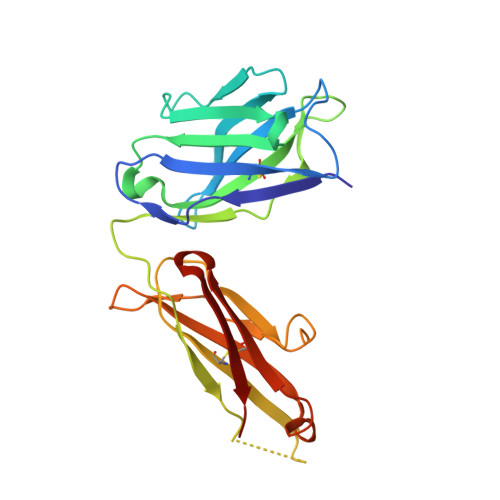
Apo2L/TRAIL and the Death Receptor 5 Agonist Antibody AMG 655 Cooperate to Promote Receptor Clustering and Antitumor Activity.
Graves, J.D., Kordich, J.J., Huang, T.H., Piasecki, J., Bush, T.L., Sullivan, T., Foltz, I.N., Chang, W., Douangpanya, H., Dang, T., O'Neill, J.W., Mallari, R., Zhao, X., Branstetter, D.G., Rossi, J.M., Long, A.M., Huang, X., Holland, P.M.(2014) Cancer Cell 26: 177-189
- PubMed: 25043603
- DOI: https://doi.org/10.1016/j.ccr.2014.04.028
- Primary Citation of Related Structures:
4N90 - PubMed Abstract:
Death receptor agonist therapies have exhibited limited clinical benefit to date. Investigations into why Apo2L/TRAIL and AMG 655 preclinical data were not predictive of clinical response revealed that coadministration of Apo2L/TRAIL with AMG 655 leads to increased antitumor activity in vitro and in vivo. The combination of Apo2L/TRAIL and AMG 655 results in enhanced signaling and can sensitize Apo2L/TRAIL-resistant cells. Structure determination of the Apo2L/TRAIL-DR5-AMG 655 ternary complex illustrates how higher order clustering of DR5 is achieved when both agents are combined. Enhanced agonism generated by combining Apo2L/TRAIL and AMG 655 provides insight into the limited efficacy observed in previous clinical trials and suggests testable hypotheses to reconsider death receptor agonism as a therapeutic strategy.
- Department of Oncology Research, Amgen Inc., Seattle, WA 98119, USA.
Organizational Affiliation: