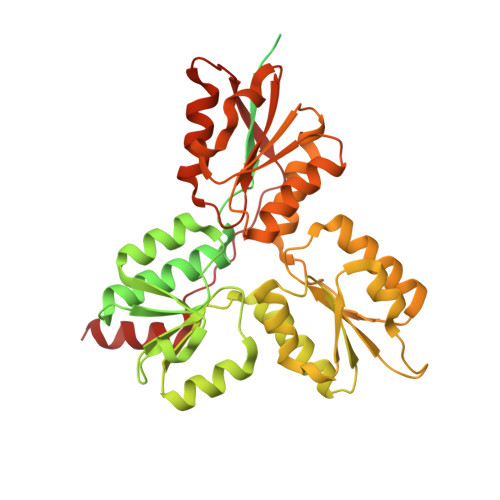
Structure of the (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase from Plasmodium falciparum.
Rekittke, I., Olkhova, E., Wiesner, J., Demmer, U., Warkentin, E., Jomaa, H., Ermler, U.(2013) FEBS Lett 587: 3968-3972
- PubMed: 24188825
- DOI: https://doi.org/10.1016/j.febslet.2013.10.029
- Primary Citation of Related Structures:
4N7B - PubMed Abstract:
Terpenoid precursor biosynthesis occurs in human and many pathogenic organisms via the mevalonate and 2-C-methyl-d-erythritol-4-phosphate (MEP) pathways, respectively. We determined the X-ray structure of the Fe/S containing (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase (LytB) of the pathogenic protozoa Plasmodium falciparum which catalyzes the terminal step of the MEP pathway. The cloverleaf fold and the active site of P. falciparum LytB corresponds to those of the Aquifex aeolicus and Escherichia coli enzymes. Its distinct electron donor [2Fe-2S] ferredoxin was modeled to its binding site by docking calculations. The presented structural data provide a platform for a rational search of anti-malarian drugs.
- Medizinische Klinik IV (Hämatologie), Justus-Liebig-Universität Giessen, Klinikstrasse 33, D-35392 Giessen, Germany.
Organizational Affiliation: