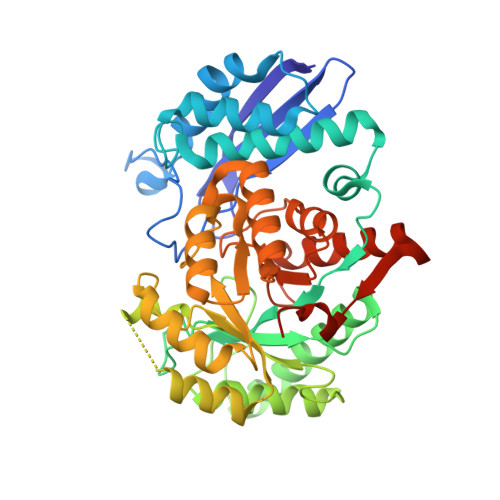
Crystal structure of an efficacious gonococcal adherence inhibitor: An enolase from Lactobacillus gasseri.
Raghunathan, K., Harris, P.T., Spurbeck, R.R., Arvidson, C.G., Arvidson, D.N.(2014) FEBS Lett 588: 2212-2216
- PubMed: 24859038
- DOI: https://doi.org/10.1016/j.febslet.2014.05.020
- Primary Citation of Related Structures:
4MKS - PubMed Abstract:
Enolases are highly conserved metalloenzymes ubiquitous to cellular metabolism. While these enzymes share a large degree of sequence and structural similarity, they have been shown to possess a wide range of moonlighting functions. Recent studies showed that an enolase from Lactobacillus gasseri impedes the ability of Neisseria gonorrhoeae to adhere to epithelial cells. We present the crystal structure of this enolase, the first from Lactobacillus, with one of its Mg(2+) cofactors. Determined using molecular replacement to 2.08Å, the structure has a flexible and surface exposed catalytic loop containing lysines, and may play a role in the inhibitory function.
- Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI 48824, USA. Electronic address: kannan@pa.msu.edu.
Organizational Affiliation: