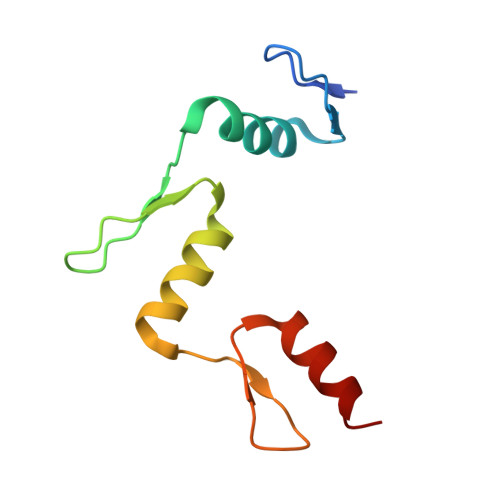
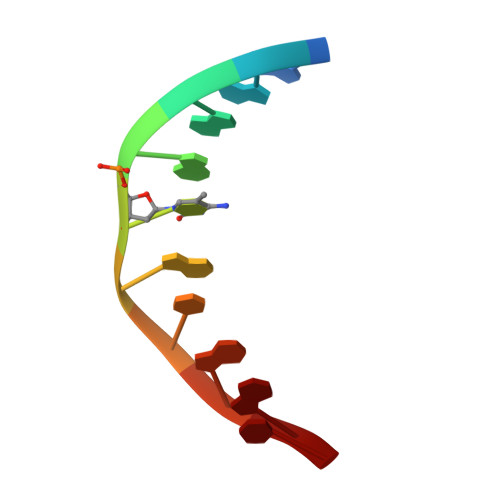
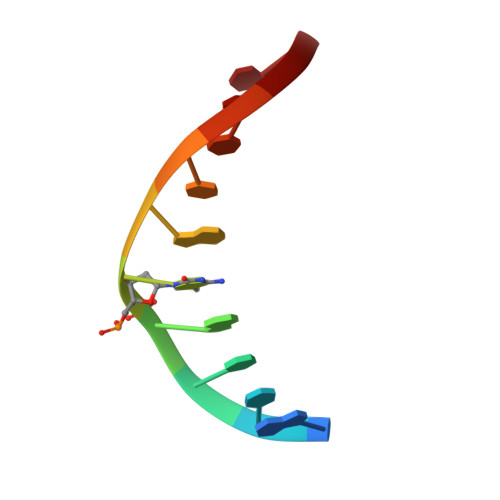
Structural basis for Klf4 recognition of methylated DNA.
Liu, Y., Olanrewaju, Y.O., Zheng, Y., Hashimoto, H., Blumenthal, R.M., Zhang, X., Cheng, X.(2014) Nucleic Acids Res 42: 4859-4867
- PubMed: 24520114
- DOI: https://doi.org/10.1093/nar/gku134
- Primary Citation of Related Structures:
4M9E - PubMed Abstract:
Transcription factor Krüppel-like factor 4 (Klf4), one of the factors directing cellular reprogramming, recognizes the CpG dinucleotide (whether methylated or unmodified) within a specific G/C-rich sequence. The binding affinity of the mouse Klf4 DNA-binding domain for methylated DNA is only slightly stronger than that for an unmodified oligonucleotide. The structure of the C-terminal three Krüppel-like zinc fingers (ZnFs) of mouse Klf4, in complex with fully methylated DNA, was determined at 1.85 Å resolution. An arginine and a glutamate interact with the methyl group. By comparison with two other recently characterized structures of ZnF protein complexes with methylated DNA, we propose a common principle of recognition of methylated CpG by C2H2 ZnF proteins, which involves a spatially conserved Arg-Glu pair.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA, New England Biolabs, 240 County Road, Ipswich, MA 01938, USA and Department of Medical Microbiology and Immunology and Program in Bioinformatics, The University of Toledo College of Medicine and Life Sciences, Toledo, OH 43614, USA.
Organizational Affiliation: