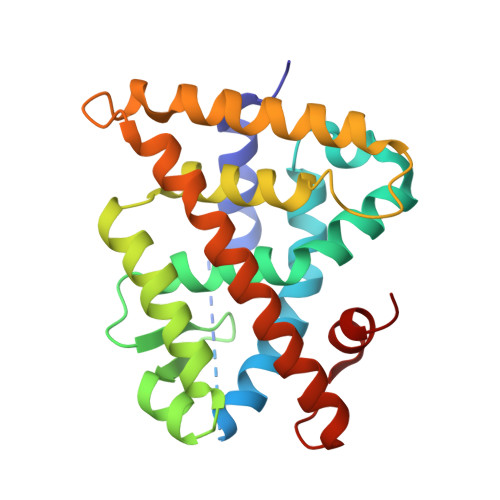
Methyl-substituted conformationally constrained rexinoid agonists for the retinoid X receptors demonstrate improved efficacy for cancer therapy and prevention.
Desphande, A., Xia, G., Boerma, L.J., Vines, K.K., Atigadda, V.R., Lobo-Ruppert, S., Grubbs, C.J., Moeinpour, F.L., Smith, C.D., Christov, K., Brouillette, W.J., Muccio, D.D.(2014) Bioorg Med Chem 22: 178-185
- PubMed: 24359708
- DOI: https://doi.org/10.1016/j.bmc.2013.11.039
- Primary Citation of Related Structures:
4M8E, 4M8H - PubMed Abstract:
(2E,4E,6Z,8Z)-8-(3',4'-Dihydro-1'(2H)-naphthalen-1'-ylidene)-3,7-dimethyl-2,3,6-octatrienoinic acid, 9cUAB30, is a selective rexinoid for the retinoid X nuclear receptors (RXR). 9cUAB30 displays substantial chemopreventive capacity with little toxicity and is being translated to the clinic as a novel cancer prevention agent. To improve on the potency of 9cUAB30, we synthesized 4-methyl analogs of 9cUAB30, which introduced chirality at the 4-position of the tetralone ring. The syntheses and biological evaluations of the racemic homolog and enantiomers are reported. We demonstrate that the S-enantiomer is the most potent and least toxic even though these enantiomers bind in a similar conformation in the ligand binding domain of RXR.
- Department of Chemistry, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Organizational Affiliation: