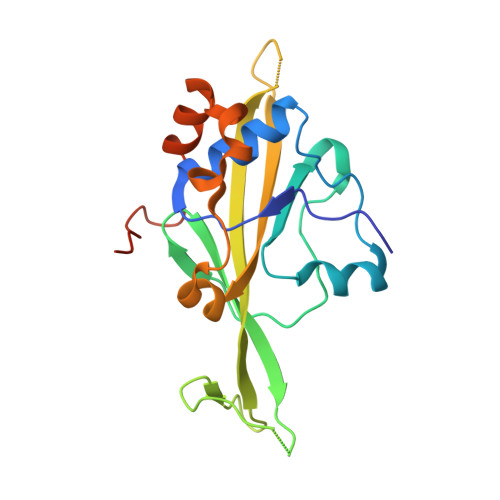
Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi.
Lohse, M.B., Rosenberg, O.S., Cox, J.S., Stroud, R.M., Finer-Moore, J.S., Johnson, A.D.(2014) Proc Natl Acad Sci U S A 111: 10404-10410
- PubMed: 24994900
- DOI: https://doi.org/10.1073/pnas.1410110111
- Primary Citation of Related Structures:
4M8B - PubMed Abstract:
WOPR-domain proteins are found throughout the fungal kingdom where they function as master regulators of cell morphology and pathogenesis. Genetic and biochemical experiments previously demonstrated that these proteins bind to specific DNA sequences and thereby regulate transcription. However, their primary sequence showed no relationship to any known DNA-binding domain, and the basis for their ability to recognize DNA sequences remained unknown. Here, we describe the 2.6-Å crystal structure of a WOPR domain in complex with its preferred DNA sequence. The structure reveals that two highly conserved regions, separated by an unconserved linker, form an interdigitated β-sheet that is tilted into the major groove of DNA. Although the main interaction surface is in the major groove, the highest-affinity interactions occur in the minor groove, primarily through a deeply penetrating arginine residue. The structure reveals a new, unanticipated mechanism by which proteins can recognize specific sequences of DNA.
- Departments of Microbiology and Immunology and.
Organizational Affiliation: