sequence specific RNA recognition of ETR3 RRM 1-2 domains
Bhatt, H., Manickam, Y., Bhavesh, N.S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CUGBP Elav-like family member 2 | B [auth A] | 180 | Homo sapiens | Mutation(s): 0 Gene Names: CELF2, BRUNOL3, CUGBP2, ETR3, NAPOR | 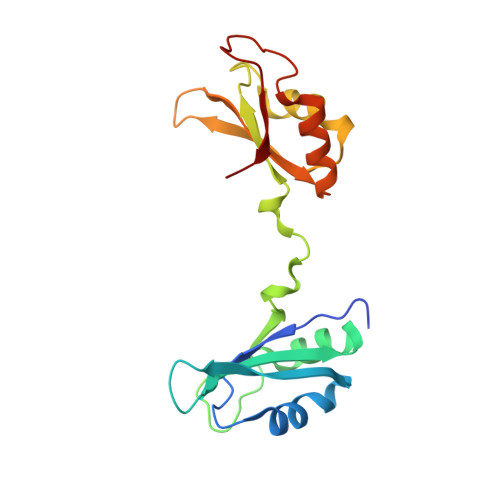 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O95319 (Homo sapiens) Explore O95319 Go to UniProtKB: O95319 | |||||
PHAROS: O95319 GTEx: ENSG00000048740 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O95319 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(*GP*CP*UP*GP*CP*GP*UP*AP*UP*UP*GP*UP*UP*UP*G)-3') | A [auth B] | 15 | N/A | 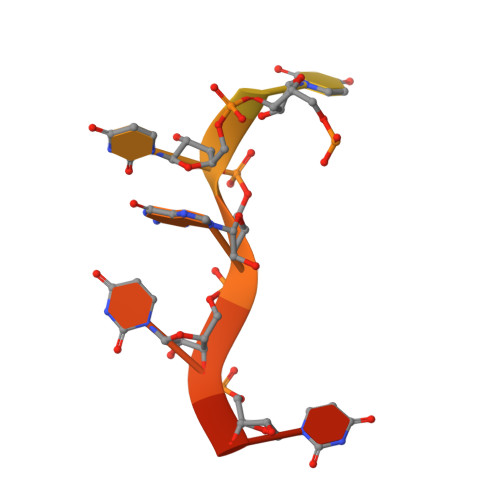 | |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 47.006 | α = 90 |
| b = 70.134 | β = 90 |
| c = 133.064 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| CCP4 | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CCP4 | phasing |