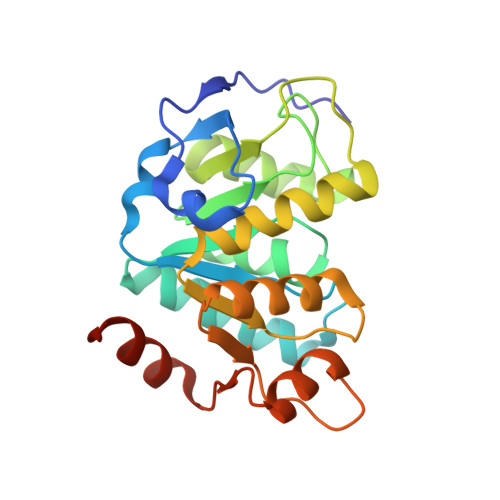
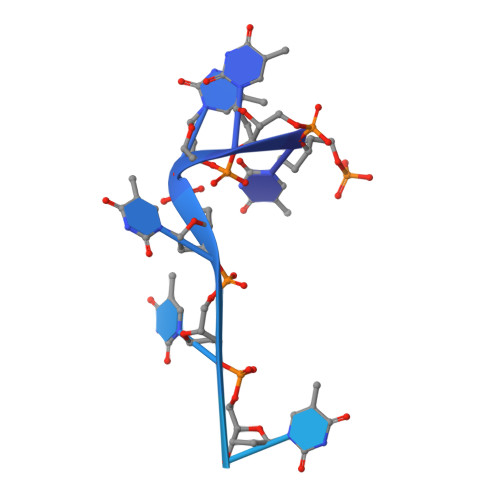
Structural insights into the unique single-stranded DNA-binding mode of Helicobacter pylori DprA.
Wang, W., Ding, J., Zhang, Y., Hu, Y., Wang, D.C.(2014) Nucleic Acids Res 42: 3478-3491
- PubMed: 24369431
- DOI: https://doi.org/10.1093/nar/gkt1334
- Primary Citation of Related Structures:
4LJK, 4LJL, 4LJR - PubMed Abstract:
Natural transformation (NT) in bacteria is a complex process, including binding, uptake, transport and recombination of exogenous DNA into the chromosome, consequently generating genetic diversity and driving evolution. DNA processing protein A (DprA), which is distributed among virtually all bacterial species, is involved in binding to the internalized single-stranded DNA (ssDNA) and promoting the loading of RecA on ssDNA during NTs. Here we present the structures of DNA_processg_A (DprA) domain of the Helicobacter pylori DprA (HpDprA) and its complex with an ssDNA at 2.20 and 1.80 Å resolutions, respectively. The complex structure revealed for the first time how the conserved DprA domain binds to ssDNA. Based on structural comparisons and binding assays, a unique ssDNA-binding mode is proposed: the dimer of HpDprA binds to ssDNA through two small, positively charged binding pockets of the DprA domains with classical Rossmann folds and the key residue Arg52 is re-oriented to 'open' the pocket in order to accommodate one of the bases of ssDNA, thus enabling HpDprA to grasp substrate with high affinity. This mode is consistent with the oligomeric composition of the complex as shown by electrophoretic mobility-shift assays and static light scattering measurements, but differs from the direct polymeric complex of Streptococcus pneumoniae DprA-ssDNA.
- The National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing 100101, China.
Organizational Affiliation: