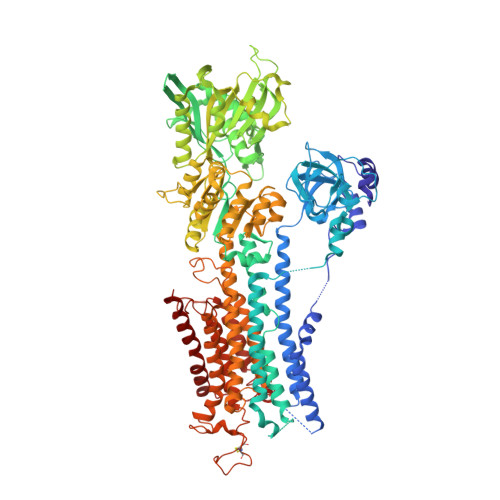
The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
Akin, B.L., Hurley, T.D., Chen, Z., Jones, L.R.(2013) J Biological Chem 288: 30181-30191
- PubMed: 23996003
- DOI: https://doi.org/10.1074/jbc.M113.501585
- Primary Citation of Related Structures:
4KYT, 4Y3U - PubMed Abstract:
P-type ATPases are a large family of enzymes that actively transport ions across biological membranes by interconverting between high (E1) and low (E2) ion-affinity states; these transmembrane transporters carry out critical processes in nearly all forms of life. In striated muscle, the archetype P-type ATPase, SERCA (sarco(endo)plasmic reticulum Ca(2+)-ATPase), pumps contractile-dependent Ca(2+) ions into the lumen of sarcoplasmic reticulum, which initiates myocyte relaxation and refills the sarcoplasmic reticulum in preparation for the next contraction. In cardiac muscle, SERCA is regulated by phospholamban (PLB), a small inhibitory phosphoprotein that decreases the Ca(2+) affinity of SERCA and attenuates contractile strength. cAMP-dependent phosphorylation of PLB reverses Ca(2+)-ATPase inhibition with powerful contractile effects. Here we present the long sought crystal structure of the PLB-SERCA complex at 2.8-Å resolution. The structure was solved in the absence of Ca(2+) in a novel detergent system employing alkyl mannosides. The structure shows PLB bound to a previously undescribed conformation of SERCA in which the Ca(2+) binding sites are collapsed and devoid of divalent cations (E2-PLB). This new structure represents one of the key unsolved conformational states of SERCA and provides a structural explanation for how dephosphorylated PLB decreases Ca(2+) affinity and depresses cardiac contractility.
- From the Krannert Institute of Cardiology and the Departments of Medicine and. Electronic address: bbudryns@iu.edu.
Organizational Affiliation: