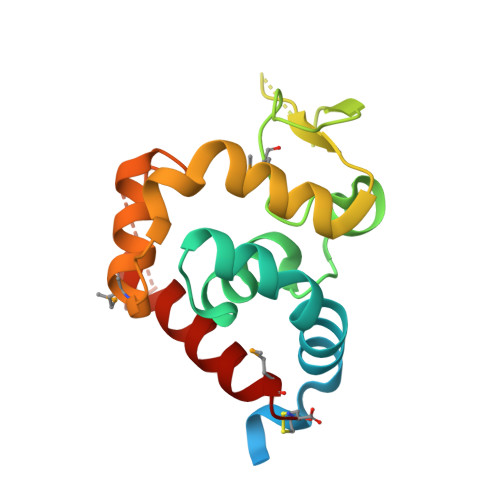
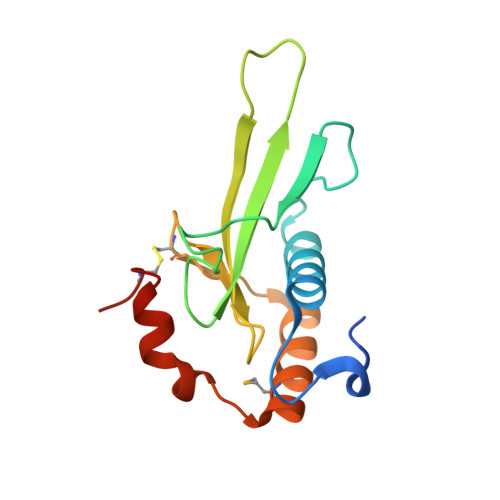
Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair.
Whitney, J.C., Chou, S., Russell, A.B., Biboy, J., Gardiner, T.E., Ferrin, M.A., Brittnacher, M., Vollmer, W., Mougous, J.D.(2013) J Biological Chem 288: 26616-26624
- PubMed: 23878199
- DOI: https://doi.org/10.1074/jbc.M113.488320
- Primary Citation of Related Structures:
4KT3 - PubMed Abstract:
Bacteria employ type VI secretion systems (T6SSs) to facilitate interactions with prokaryotic and eukaryotic cells. Despite the widespread identification of T6SSs among Gram-negative bacteria, the number of experimentally validated substrate effector proteins mediating these interactions remains small. Here, employing an informatics approach, we define novel families of T6S peptidoglycan glycoside hydrolase effectors. Consistent with the known intercellular self-intoxication exhibited by the T6S pathway, we observe that each effector gene is located adjacent to a hypothetical open reading frame encoding a putative periplasmically localized immunity determinant. To validate our sequence-based approach, we functionally investigate a representative family member from the soil-dwelling bacterium Pseudomonas protegens. We demonstrate that this protein is secreted in a T6SS-dependent manner and that it confers a fitness advantage in growth competition assays with Pseudomonas putida. In addition, we determined the 1.4 Å x-ray crystal structure of this effector in complex with its cognate immunity protein. The structure reveals the effector shares highest overall structural similarity to a glycoside hydrolase family associated with peptidoglycan N-acetylglucosaminidase activity, suggesting that T6S peptidoglycan glycoside hydrolase effector families may comprise significant enzymatic diversity. Our structural analyses also demonstrate that self-intoxication is prevented by the immunity protein through direct occlusion of the effector active site. This work significantly expands our current understanding of T6S effector diversity.
- From the Departments of Microbiology and.
Organizational Affiliation: