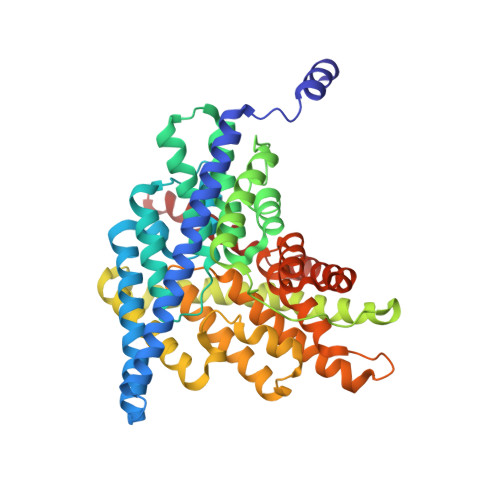
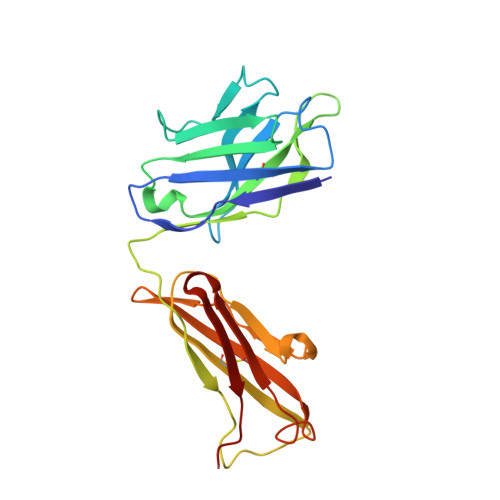
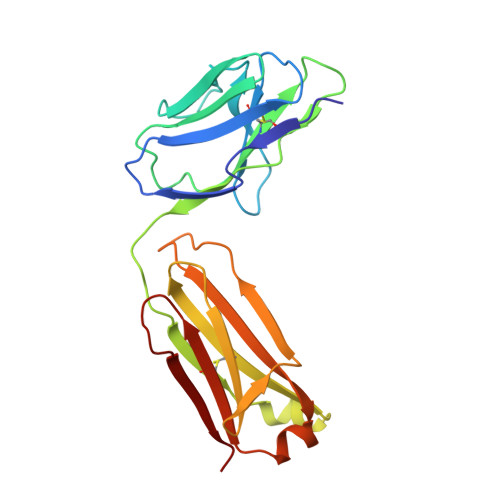
Fluoride-dependent interruption of the transport cycle of a CLC Cl(-)/H(+) antiporter.
Lim, H.H., Stockbridge, R.B., Miller, C.(2013) Nat Chem Biol 9: 721-725
- PubMed: 24036509
- DOI: https://doi.org/10.1038/nchembio.1336
- Primary Citation of Related Structures:
4KJP, 4KJQ, 4KJW, 4KK5, 4KK6, 4KK8, 4KK9, 4KKA, 4KKB, 4KKC, 4KKL - PubMed Abstract:
Cl(-)/H(+) antiporters of the CLC superfamily transport anions across biological membranes in varied physiological contexts. These proteins are weakly selective among anions commonly studied, including Cl(-), Br(-), I(-), NO3(-) and SCN(-), but they seem to be very selective against F(-). The recent discovery of a new CLC clade of F(-)/H(+) antiporters, which are highly selective for F(-) over Cl(-), led us to investigate the mechanism of Cl(-)-over-F(-) selectivity by a CLC Cl(-)/H(+) antiporter, CLC-ec1. By subjecting purified CLC-ec1 to anion transport measurements, electrophysiological recording, equilibrium ligand-binding studies and X-ray crystallography, we show that F(-) binds in the Cl(-) transport pathway with affinity similar to Cl(-) but stalls the transport cycle. Examination of various mutant antiporters implies a 'lock-down' mechanism of F(-) inhibition, in which F(-), by virtue of its unique hydrogen-bonding chemistry, greatly retards a proton-linked conformational change essential for the transport cycle of CLC-ec1.
- 1] Department of Biochemistry, Brandeis University, Waltham, Massachusetts, USA. [2] Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts, USA. [3].
Organizational Affiliation: