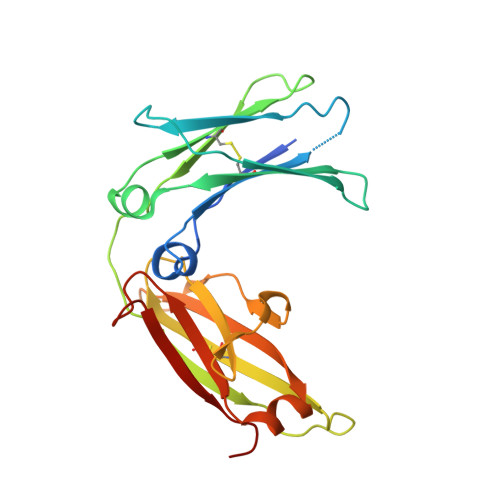
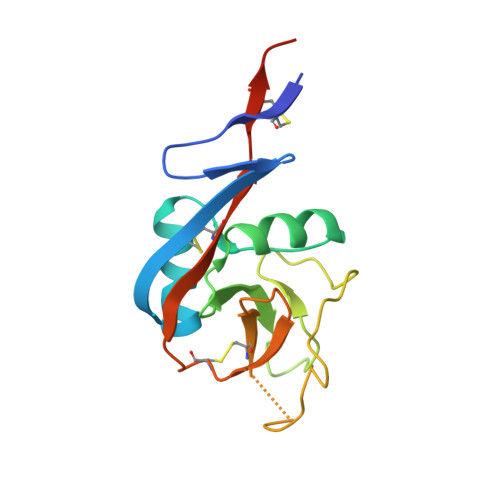
A range of C3-C4 interdomain angles in IgE Fc accommodate binding to its receptor CD23.
Dhaliwal, B., Pang, M.O., Yuan, D., Beavil, A.J., Sutton, B.J.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 305-309
- PubMed: 24598915
- DOI: https://doi.org/10.1107/S2053230X14003355
- Primary Citation of Related Structures:
4KI1 - PubMed Abstract:
The antibody IgE plays a central role in allergic disease, functioning principally through two cell-surface receptors: FcℇRI and CD23. FcℇRI on mast cells and basophils mediates the immediate hypersensitivity response, whilst the interaction of IgE with CD23 on B cells regulates IgE production. Crystal structures of the lectin-like `head' domain of CD23 alone and bound to a subfragment of IgE consisting of the dimer of Cℇ3 and Cℇ4 domains (Fcℇ3-4) have recently been determined, revealing flexibility in the IgE-binding site of CD23. Here, a new crystal form of the CD23-Fcℇ3-4 complex with different molecular-packing constraints is reported, which together with the earlier results demonstrates that conformational variability at the interface extends additionally to the IgE Fc and the quaternary structure of its domains.
- Randall Division of Cell and Molecular Biophysics, King's College London, New Hunt's House, Guy's Campus, London SE1 1UL, England.
Organizational Affiliation: