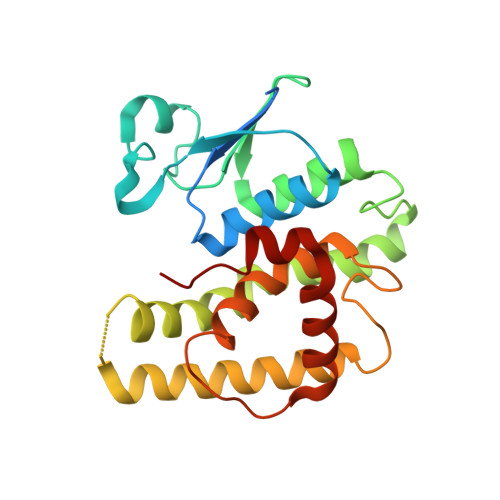
Crystal structure of a glutathione transferase family member from Salmonella enterica ty2, target efi-507262, with bound glutathione
Vetting, M.W., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Stead, M., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Gerlt, J.A., Armstrong, R.N., Almo, S.C., Enzyme Function Initiative (EFI)To be published.