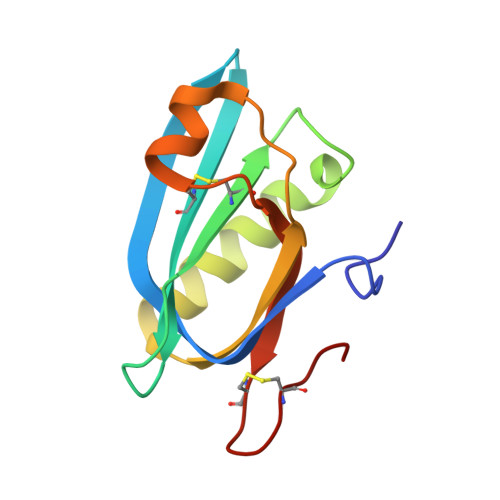
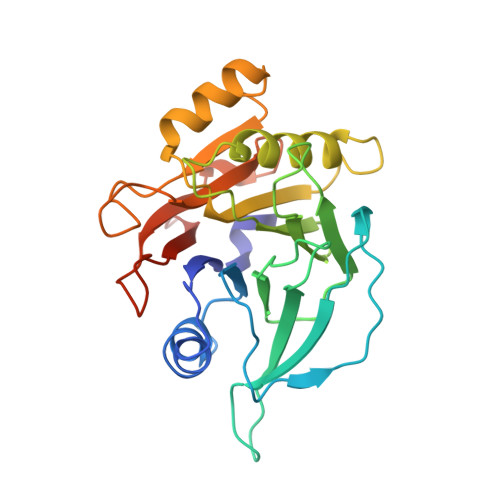
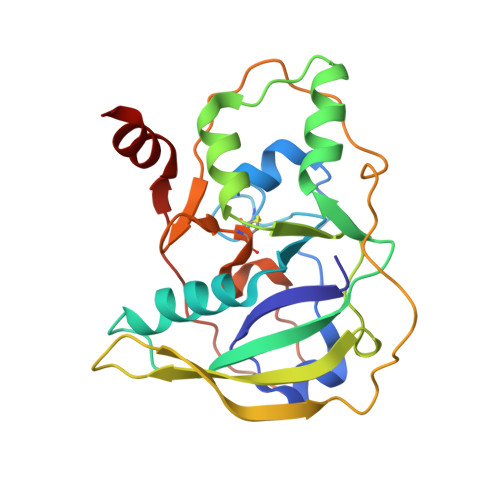
Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin.
Song, J., Gao, X., Galan, J.E.(2013) Nature 499: 350-354
- PubMed: 23842500
- DOI: https://doi.org/10.1038/nature12377
- Primary Citation of Related Structures:
4K6L - PubMed Abstract:
Salmonella enterica serovar Typhi (S. Typhi) differs from most other salmonellae in that it causes a life-threatening systemic infection known as typhoid fever. The molecular bases for its unique clinical presentation are unknown. Here we find that the systemic administration of typhoid toxin, a unique virulence factor of S. Typhi, reproduces many of the acute symptoms of typhoid fever in an animal model. We identify specific carbohydrate moieties on specific surface glycoproteins that serve as receptors for typhoid toxin, which explains its broad cell target specificity. We present the atomic structure of typhoid toxin, which shows an unprecedented A2B5 organization with two covalently linked A subunits non-covalently associated to a pentameric B subunit. The structure provides insight into the toxin's receptor-binding specificity and delivery mechanisms and reveals how the activities of two powerful toxins have been co-opted into a single, unique toxin that can induce many of the symptoms characteristic of typhoid fever. These findings may lead to the development of potentially life-saving therapeutics against typhoid fever.
- Department of Microbial Pathogenesis, Yale University School of Medicine, New Haven, Connecticut 06536, USA.
Organizational Affiliation: