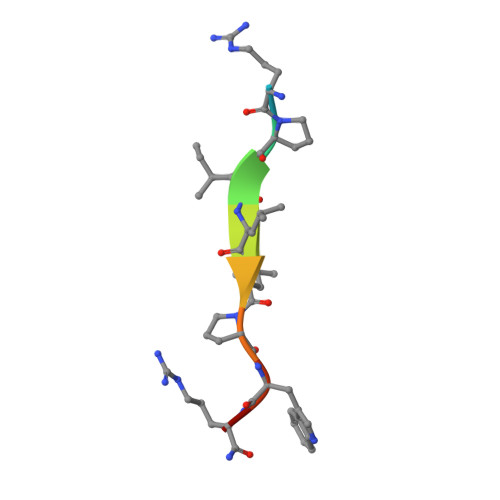
Structural Identification of DnaK Binding Sites within Bovine and Sheep Bactenecin Bac7.
Zahn, M., Kieslich, B., Berthold, N., Knappe, D., Hoffmann, R., Strater, N.(2014) Protein Pept Lett 21: 407-412
- PubMed: 24164259
- DOI: https://doi.org/10.2174/09298665113206660111
- Primary Citation of Related Structures:
4JWC, 4JWD, 4JWE, 4JWI - PubMed Abstract:
Bacterial resistance against common antibiotics is an increasing health problem. New pharmaceuticals for the treatment of infections caused by resistant pathogens are needed. Small proline-rich antimicrobial peptides (PrAMPs) from insects are known to bind intracellularly to the conventional substrate binding cleft of the E. coli Hsp70 chaperone DnaK. Furthermore, bactenecins from mammals, members of the cathelicidin family, also contain potential DnaK binding sites. Crystal structures of bovine and sheep Bac7 in complex with the DnaK substrate binding domain show that the peptides bind in the forward binding mode with a leucine positioned in the central hydrophobic pocket. In most structures, proline and arginine residues preceding leucine occupy the hydrophobic DnaK binding sites -1 and -2. Within bovine Bac7, four potential DnaK binding sites were identified.
- Institute of Bioanalytical Chemistry, Center for Biotechnology and Biomedicine, University of Leipzig, Deutscher Platz 5, 04103 Leipzig, Germany. strater@bbz.uni-leipzig.de.
Organizational Affiliation: