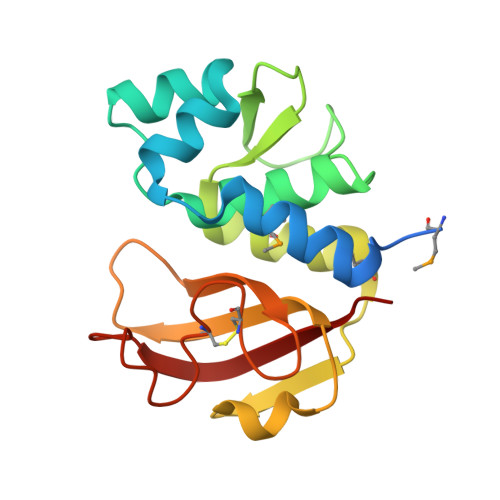
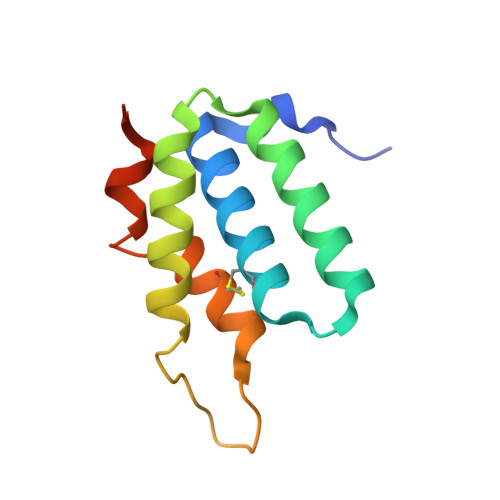
Insights into the cross-immunity mechanism within effector families of bacteria type VI secretion system from the structure of StTae4-EcTai4 complex
Zhang, H., Gao, Z.Q., Wei, Y., Xu, J.H., Dong, Y.H.(2013) PLoS One 8: e73782-e73782
- PubMed: 24023903
- DOI: https://doi.org/10.1371/journal.pone.0073782
- Primary Citation of Related Structures:
4JUR - PubMed Abstract:
The Gram-negative bacteria type VI secretion system (T6SS) has been found to play an important role in interbacterial competition, biofilm formation and many other virulence-related processes. The bacteria harboring T6SS inject the effectors into their recipient's cytoplasm or periplasm to kill them and meanwhile, to avoid inhibiting itself, the cognate immunity proteins were produced to acts as the effector inhibitor. Tae4 (type VI amidase effector 4) and Tai4 (type VI amidase immunity 4) are newly identified T6SS effector-immunity (EI) pairs. We have recently solved the structures of StTae4-Tai4 and EcTae4-Tai4 complexes from the human pathogens Salmonella typhimurium and Enterobacter cloacae, respectively. It is very interesting and important to discover whether there is cross-neutralization between St- and EcTai4 and whether their effector inhibition mechanism is conserved. Here, we determined the crystal structure of StTae4 in complex with EcTai4. The solution conformation study revealed it is a compact heterotetramer that consists of an EcTai4 homodimer binding two StTae4 molecules in solution, different from that in crystal. A remarkable shift can be observed in both the flexible winding loop of StTae4 and protruding loop of EcTai4 and disulfide bonds are formed to stabilize their overall conformations. The in vitro and in vivo interactions studies showed EcTai4 can efficiently rescue the cells from the toxicity of its cognate effectors StTae4, but can not neutralize the toxic activities of the effectors from other families. These findings provide clear structural evidence to support the previous observation of cross-immunity within T6SS families and provide a basis for understanding their important roles in polymicrobial environments.
- Beijing Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences, Beijing, People's Republic of China.
Organizational Affiliation: