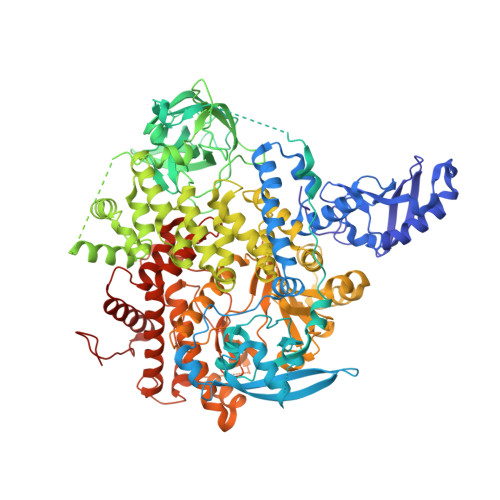
Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation.
Furet, P., Guagnano, V., Fairhurst, R.A., Imbach-Weese, P., Bruce, I., Knapp, M., Fritsch, C., Blasco, F., Blanz, J., Aichholz, R., Hamon, J., Fabbro, D., Caravatti, G.(2013) Bioorg Med Chem Lett 23: 3741-3748
- PubMed: 23726034
- DOI: https://doi.org/10.1016/j.bmcl.2013.05.007
- Primary Citation of Related Structures:
4JPS - PubMed Abstract:
Phosphatidylinositol-3-kinase α (PI3Kα) is a therapeutic target of high interest in anticancer drug research. On the basis of a binding model rationalizing the high selectivity and potency of a particular series of 2-aminothiazole compounds in inhibiting PI3Kα, a medicinal chemistry program has led to the discovery of the clinical candidate NVP-BYL719.
- Novartis Institutes for BioMedical Research, WKL-136.4.12, CH-4002 Basel, Switzerland.
Organizational Affiliation: