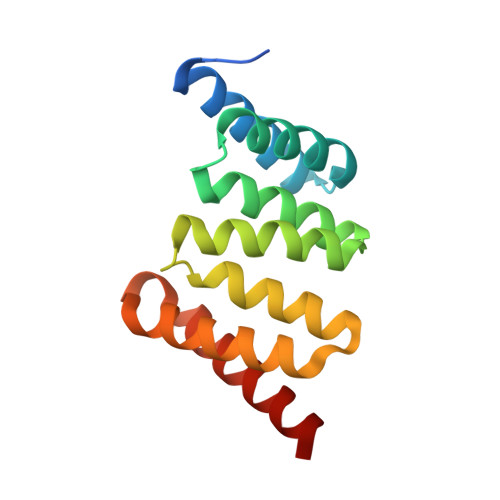
Membrane and Chaperone Recognition by the Major Translocator Protein PopB of the Type III Secretion System of Pseudomonas aeruginosa.
Discola, K.F., Forster, A., Boulay, F., Simorre, J.P., Attree, I., Dessen, A., Job, V.(2014) J Biological Chem 289: 3591-3601
- PubMed: 24297169
- DOI: https://doi.org/10.1074/jbc.M113.517920
- Primary Citation of Related Structures:
4JL0 - PubMed Abstract:
The type III secretion system is a widespread apparatus used by pathogenic bacteria to inject effectors directly into the cytoplasm of eukaryotic cells. A key component of this highly conserved system is the translocon, a pore formed in the host membrane that is essential for toxins to bypass this last physical barrier. In Pseudomonas aeruginosa the translocon is composed of PopB and PopD, both of which before secretion are stabilized within the bacterial cytoplasm by a common chaperone, PcrH. In this work we characterize PopB, the major translocator, in both membrane-associated and PcrH-bound forms. By combining sucrose gradient centrifugation experiments, limited proteolysis, one-dimensional NMR, and β-lactamase reporter assays on eukaryotic cells, we show that PopB is stably inserted into bilayers with its flexible N-terminal domain and C-terminal tail exposed to the outside. In addition, we also report the crystal structure of the complex between PcrH and an N-terminal region of PopB (residues 51-59), which reveals that PopB lies within the concave face of PcrH, employing mostly backbone residues for contact. PcrH is thus the first chaperone whose structure has been solved in complex with both type III secretion systems translocators, revealing that both molecules employ the same surface for binding and excluding the possibility of formation of a ternary complex. The characterization of the major type III secretion system translocon component in both membrane-bound and chaperone-bound forms is a key step for the eventual development of antibacterials that block translocon assembly.
- From the Institut de Biologie Structurale, Université Grenoble Alpes, 6 rue Jules Horowitz, 38000 Grenoble, France.
Organizational Affiliation: