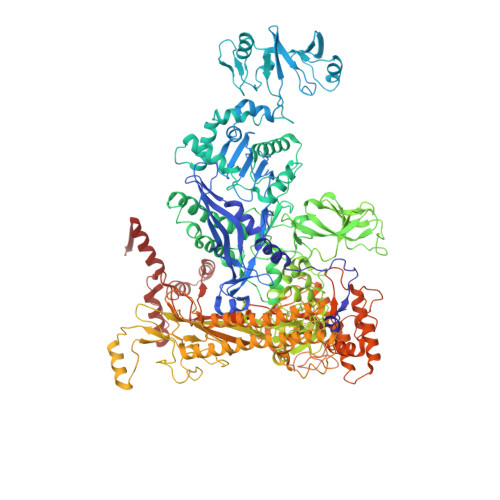
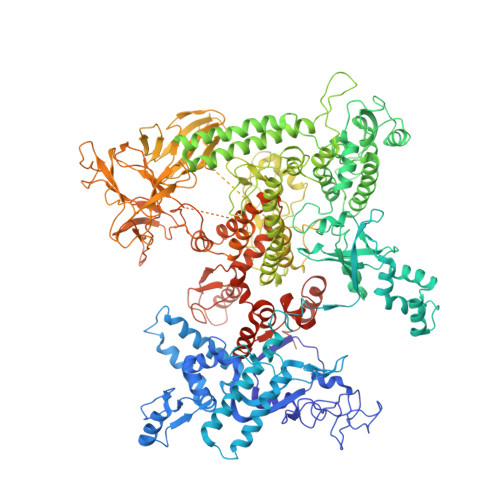
The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex.
Zuo, Y., Wang, Y., Steitz, T.A.(2013) Mol Cell 50: 430-436
- PubMed: 23623685
- DOI: https://doi.org/10.1016/j.molcel.2013.03.020
- Primary Citation of Related Structures:
4JKR - PubMed Abstract:
Guanosine tetraphosphate (ppGpp) is an alarmone that enables bacteria to adapt to their environment. It has been known for years that ppGpp acts directly on RNA polymerase (RNAP) to alter the rate of transcription, but its exact target site is still under debate. Here we report a crystal structure of Escherichia coli RNAP holoenzyme in complex with ppGpp at 4.5 Å resolution. The structure reveals that ppGpp binds at an interface between the shelf and core modules on the outer surface of RNAP, away from the catalytic center and the nucleic acid binding path. Bound ppGpp connects these two pivotal modules that may restrain the opening of the RNAP cleft. A detailed mechanism of action of ppGpp is proposed in which ppGpp prevents the closure of the active center that is induced by the binding of NTP, which could slow down nucleotide addition cycles and destabilize the initial transcription complexes.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: