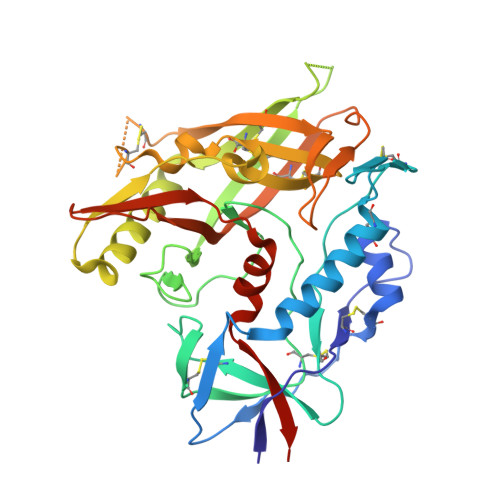
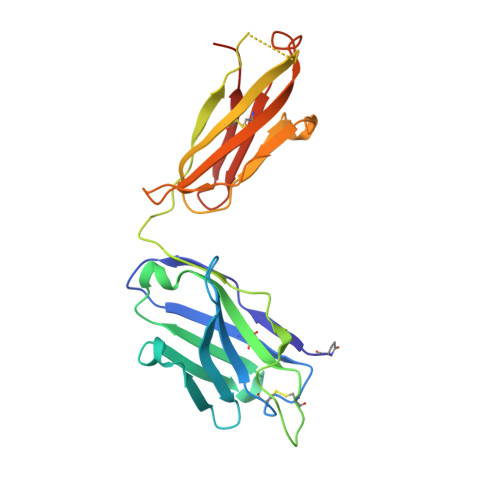
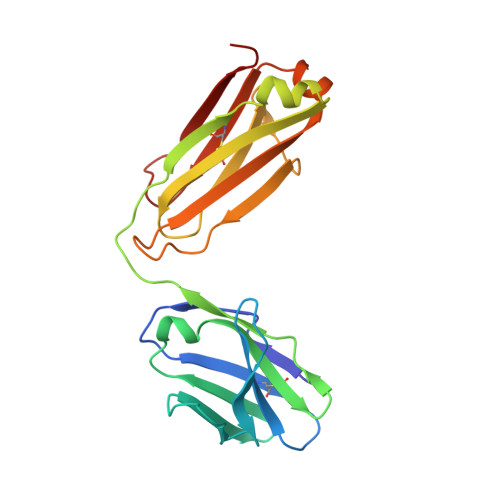
Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies.
Diskin, R., Klein, F., Horwitz, J.A., Halper-Stromberg, A., Sather, D.N., Marcovecchio, P.M., Lee, T., West, A.P., Gao, H., Seaman, M.S., Stamatatos, L., Nussenzweig, M.C., Bjorkman, P.J.(2013) J Exp Medicine 210: 1235-1249
- PubMed: 23712429
- DOI: https://doi.org/10.1084/jem.20130221
- Primary Citation of Related Structures:
4JKP - PubMed Abstract:
Recently identified broadly neutralizing antibodies (bNAbs) that potently neutralize most HIV-1 strains are key to potential antibody-based therapeutic approaches to combat HIV/AIDS in the absence of an effective vaccine. Increasing bNAb potencies and resistance to common routes of HIV-1 escape through mutation would facilitate their use as therapeutics. We previously used structure-based design to create the bNAb NIH45-46(G54W), which exhibits superior potency and/or breadth compared with other bNAbs. We report new, more effective NIH45-46(G54W) variants designed using analyses of the NIH45-46-gp120 complex structure and sequences of NIH45-46(G54W)-resistant HIV-1 strains. One variant, 45-46m2, neutralizes 96% of HIV-1 strains in a cross-clade panel and viruses isolated from an HIV-infected individual that are resistant to all other known bNAbs, making it the single most broad and potent anti-HIV-1 antibody to date. A description of its mechanism is presented based on a 45-46m2-gp120 crystal structure. A second variant, 45-46m7, designed to thwart HIV-1 resistance to NIH45-46(G54W) arising from mutations in a gp120 consensus sequence, targets a common route of HIV-1 escape. In combination, 45-46m2 and 45-46m7 reduce the possible routes for the evolution of fit viral escape mutants in HIV-1YU-2-infected humanized mice, with viremic control exhibited when a third antibody, 10-1074, was added to the combination.
- Division of Biology, California Institute of Technology, Pasadena, CA 91125, USA. ron.diskin@weizmann.ac.il
Organizational Affiliation: