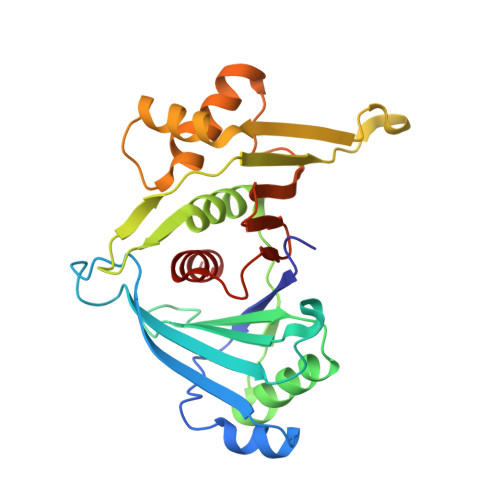
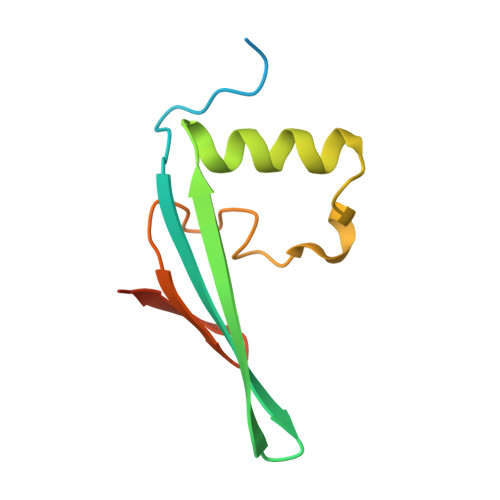
An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Hinshaw, S.M., Harrison, S.C.(2013) Cell Rep 5: 29-36
- PubMed: 24075991
- DOI: https://doi.org/10.1016/j.celrep.2013.08.036
- Primary Citation of Related Structures:
4IT3, 4JE3 - PubMed Abstract:
Accurate segregation of genetic material in eukaryotes relies on the kinetochore, a multiprotein complex that connects centromeric DNA with microtubules. In yeast and humans, two proteins-Mif2/CENP-C and Chl4/CNEP-N-interact with specialized centromeric nucleosomes and establish distinct but cross-connecting axes of chromatin-microtubule linkage. Proteins recruited by Chl4/CENP-N include a subset that regulates chromosome transmission fidelity. We show that Chl4 and a conserved member of this subset, Iml3, both from Saccharomyces cerevisiae, form a stable protein complex that interacts with Mif2 and Sgo1. We have determined the structures of an Iml3 homodimer and an Iml3-Chl4 heterodimer, which suggest a mechanism for regulating the assembly of this functional axis of the kinetochore. We propose that at the core centromere, the Chl4-Iml3 complex participates in recruiting factors, such as Sgo1, that influence sister chromatid cohesion and encourage sister kinetochore biorientation.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: