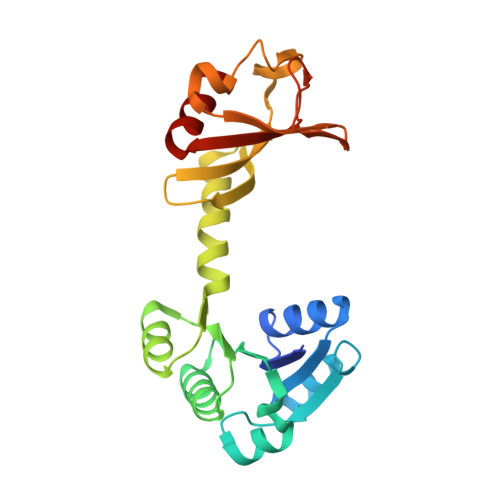
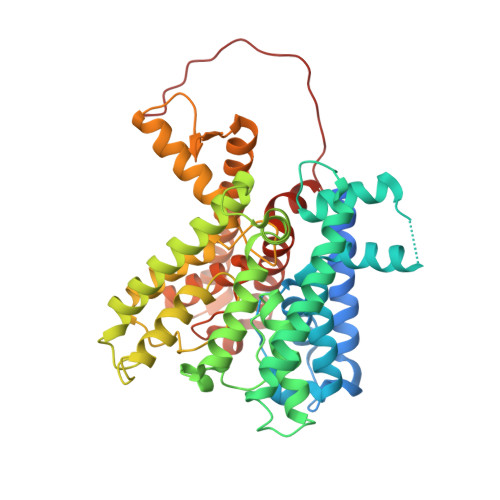
The structure of the KtrAB potassium transporter
Vieira-Pires, R.S., Szollosi, A., Morais-Cabral, J.H.(2013) Nature 496: 323-328
- PubMed: 23598340
- DOI: https://doi.org/10.1038/nature12055
- Primary Citation of Related Structures:
4J7C, 4J90, 4J91 - PubMed Abstract:
In bacteria, archaea, fungi and plants the Trk, Ktr and HKT ion transporters are key components of osmotic regulation, pH homeostasis and resistance to drought and high salinity. These ion transporters are functionally diverse: they can function as Na(+) or K(+) channels and possibly as cation/K(+) symporters. They are closely related to potassium channels both at the level of the membrane protein and at the level of the cytosolic regulatory domains. Here we describe the crystal structure of a Ktr K(+) transporter, the KtrAB complex from Bacillus subtilis. The structure shows the dimeric membrane protein KtrB assembled with a cytosolic octameric KtrA ring bound to ATP, an activating ligand. A comparison between the structure of KtrAB-ATP and the structures of the isolated full-length KtrA protein with ATP or ADP reveals a ligand-dependent conformational change in the octameric ring, raising new ideas about the mechanism of activation in these transporters.
- Instituto de Biologia Molecular e Celular, Universidade do Porto, Rua do Campo Alegre 823, Porto 4150-180, Portugal.
Organizational Affiliation: