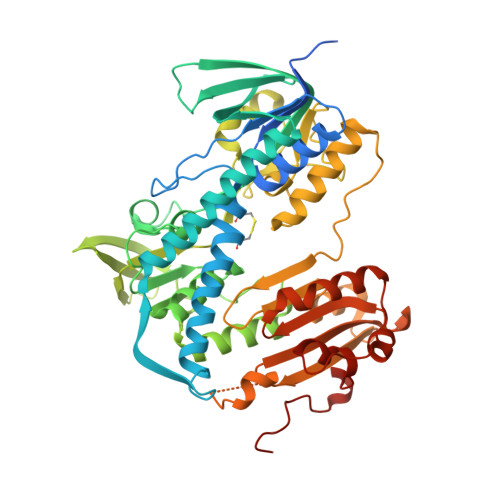
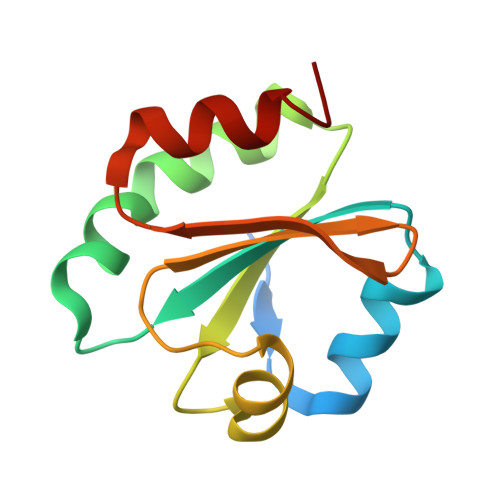
Crystal Structure of the Plasmodium falciparum Thioredoxin Reductase-Thioredoxin Complex.
Fritz-Wolf, K., Jortzik, E., Stumpf, M., Preuss, J., Iozef, R., Rahlfs, S., Becker, K.(2013) J Mol Biology 425: 3446-3460
- PubMed: 23845423
- DOI: https://doi.org/10.1016/j.jmb.2013.06.037
- Primary Citation of Related Structures:
4J56, 4J57 - PubMed Abstract:
Over the last decades, malaria parasites have been rapidly developing resistance against antimalarial drugs, which underlines the need for novel drug targets. Thioredoxin reductase (TrxR) is crucially involved in redox homeostasis and essential for Plasmodium falciparum. Here, we report the first crystal structure of P. falciparum TrxR bound to its substrate thioredoxin 1. Upon complex formation, the flexible C-terminal arm and an insertion loop of PfTrxR are rearranged, suggesting that the C-terminal arm changes its conformation during catalysis similar to human TrxR. Striking differences between P. falciparum and human TrxR are a Plasmodium-specific insertion and the conformation of the C-terminal arm, which lead to considerable differences in thioredoxin binding and disulfide reduction. Moreover, we functionally analyzed amino acid residues involved in substrate binding and in the architecture of the intersubunit cavity, which is a known binding site for disulfide reductase inhibitors. Cell biological experiments indicate that P. falciparum TrxR is indeed targeted in the parasite by specific inhibitors with antimalarial activity. Differences between P. falciparum and human TrxR and details on substrate reduction and inhibitor binding provide the first solid basis for structure-based drug development and lead optimization.
- Interdisciplinary Research Center, Justus Liebig University, D-35392 Giessen, Germany.
Organizational Affiliation: