Covalent modifications of the active site cysteine occur as a result of mutating the glutamate of the catalytic triad in the amidase from Nesterenkonia sp.
Kimani, S.W., Hunter, R., Vlok, M., Watermeyer, J., Sewell, B.T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Amidase | 283 | Nesterenkonia sp. 10004 | Mutation(s): 1 Gene Names: Nit2 EC: 3.5.1.4 | 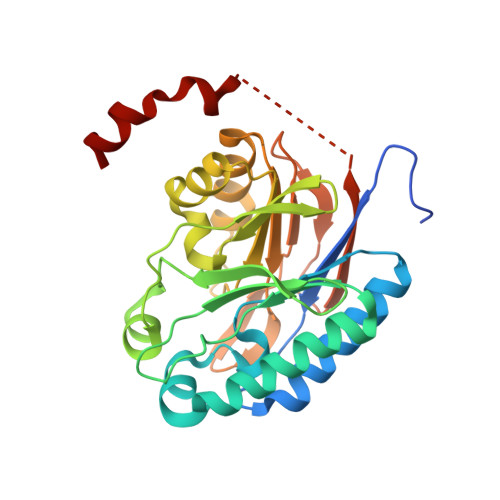 | |
UniProt | |||||
Find proteins for D0VWZ1 (Nesterenkonia sp. 10004) Explore D0VWZ1 Go to UniProtKB: D0VWZ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D0VWZ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | B [auth A], C [auth A], D [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| BMD Query on BMD | E [auth A] | BUTYRAMIDE C4 H9 N O DNSISZSEWVHGLH-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.32 | α = 90 |
| b = 114.83 | β = 90 |
| c = 65.06 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |