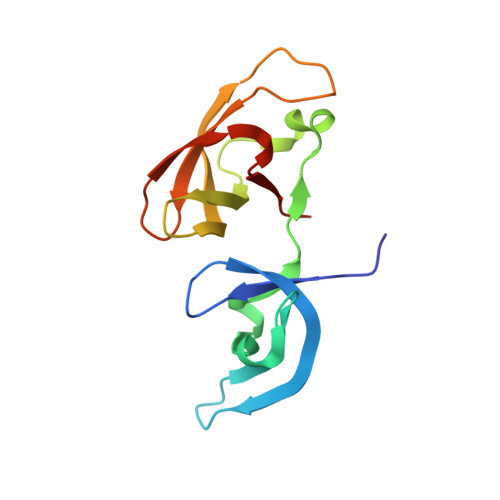
Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.
Law, J.A., Du, J., Hale, C.J., Feng, S., Krajewski, K., Palanca, A.M., Strahl, B.D., Patel, D.J., Jacobsen, S.E.(2013) Nature 498: 385-389
- PubMed: 23636332
- DOI: https://doi.org/10.1038/nature12178
- Primary Citation of Related Structures:
4IUP, 4IUQ, 4IUR, 4IUT, 4IUU, 4IUV - PubMed Abstract:
DNA methylation is an epigenetic modification that has critical roles in gene silencing, development and genome integrity. In Arabidopsis, DNA methylation is established by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) and targeted by 24-nucleotide small interfering RNAs (siRNAs) through a pathway termed RNA-directed DNA methylation (RdDM). This pathway requires two plant-specific RNA polymerases: Pol-IV, which functions to initiate siRNA biogenesis, and Pol-V, which functions to generate scaffold transcripts that recruit downstream RdDM factors. To understand the mechanisms controlling Pol-IV targeting we investigated the function of SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1), a Pol-IV-interacting protein. Here we show that SHH1 acts upstream in the RdDM pathway to enable siRNA production from a large subset of the most active RdDM targets, and that SHH1 is required for Pol-IV occupancy at these same loci. We also show that the SHH1 SAWADEE domain is a novel chromatin-binding module that adopts a unique tandem Tudor-like fold and functions as a dual lysine reader, probing for both unmethylated K4 and methylated K9 modifications on the histone 3 (H3) tail. Finally, we show that key residues within both lysine-binding pockets of SHH1 are required in vivo to maintain siRNA and DNA methylation levels as well as Pol-IV occupancy at RdDM targets, demonstrating a central role for methylated H3K9 binding in SHH1 function and providing the first insights into the mechanism of Pol-IV targeting. Given the parallels between methylation systems in plants and mammals, a further understanding of this early targeting step may aid our ability to control the expression of endogenous and newly introduced genes, which has broad implications for agriculture and gene therapy.
- Department of Molecular, Cell and Developmental Biology, University of California at Los Angeles, Los Angeles, California 90095, USA.
Organizational Affiliation: