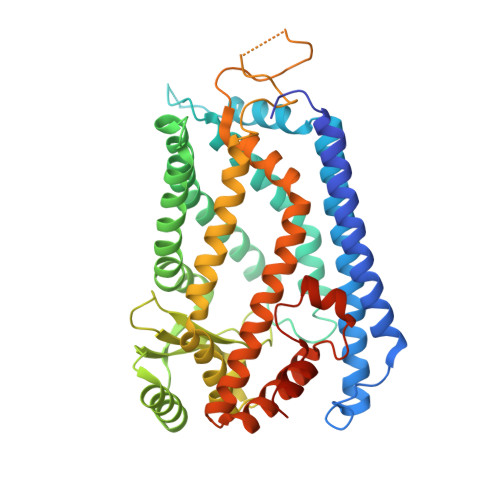
Structure of the integral membrane protein CAAX protease Ste24p.
Pryor, E.E., Horanyi, P.S., Clark, K.M., Fedoriw, N., Connelly, S.M., Koszelak-Rosenblum, M., Zhu, G., Malkowski, M.G., Wiener, M.C., Dumont, M.E.(2013) Science 339: 1600-1604
- PubMed: 23539602
- DOI: https://doi.org/10.1126/science.1232048
- Primary Citation of Related Structures:
4IL3 - PubMed Abstract:
Posttranslational lipidation provides critical modulation of the functions of some proteins. Isoprenoids (i.e., farnesyl or geranylgeranyl groups) are attached to cysteine residues in proteins containing C-terminal CAAX sequence motifs (where A is an aliphatic residue and X is any residue). Isoprenylation is followed by cleavage of the AAX amino acid residues and, in some cases, by additional proteolytic cuts. We determined the crystal structure of the CAAX protease Ste24p, a zinc metalloprotease catalyzing two proteolytic steps in the maturation of yeast mating pheromone a-factor. The Ste24p core structure is a ring of seven transmembrane helices enclosing a voluminous cavity containing the active site and substrate-binding groove. The cavity is accessible to the external milieu by means of gaps between splayed transmembrane helices. We hypothesize that cleavage proceeds by means of a processive mechanism of substrate insertion, translocation, and ejection.
- Membrane Protein Structural Biology Consortium, USA.
Organizational Affiliation: