Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
Chen, Q., Chen, X., Wang, Q., Zhang, F., Lou, Z., Zhang, Q., Zhou, D.X.(2013) PLoS Genet 9: e1003239-e1003239
- PubMed: 23357881
- DOI: https://doi.org/10.1371/journal.pgen.1003239
- Primary Citation of Related Structures:
4IGO, 4IGP, 4IGQ - PubMed Abstract:
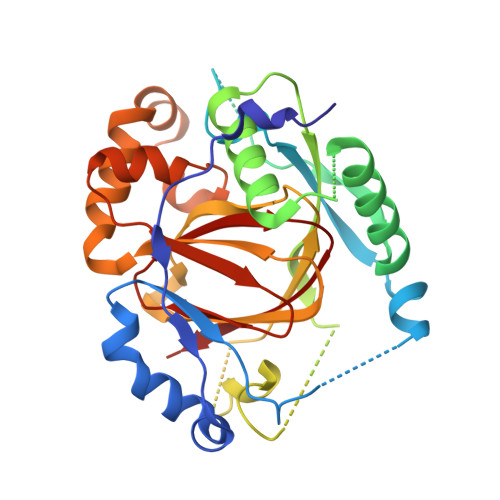
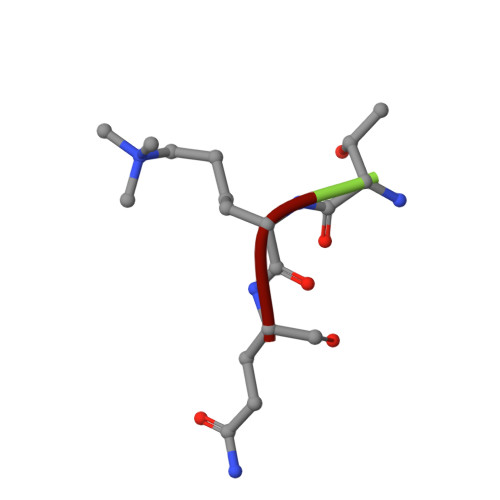
Histone lysine methylation is an important epigenetic modification in regulating chromatin structure and gene expression. Histone H3 lysine 4 methylation (H3K4me), which can be in a mono-, di-, or trimethylated state, has been shown to play an important role in gene expression involved in plant developmental control and stress adaptation. However, the resetting mechanism of this epigenetic modification is not yet fully understood. In this work, we identified a JmjC domain-containing protein, JMJ703, as a histone lysine demethylase that specifically reverses all three forms of H3K4me in rice. Loss-of-function mutation of the gene affected stem elongation and plant growth, which may be related to increased expression of cytokinin oxidase genes in the mutant. Analysis of crystal structure of the catalytic core domain (c-JMJ703) of the protein revealed a general structural similarity with mammalian and yeast JMJD2 proteins that are H3K9 and H3K36 demethylases. However, several specific features were observed in the structure of c-JMJ703. Key residues that interact with cofactors Fe(II) and N-oxalylglycine and the methylated H3K4 substrate peptide were identified and were shown to be essential for the demethylase activity in vivo. Several key residues are specifically conserved in known H3K4 demethylases, suggesting that they may be involved in the specificity for H3K4 demethylation.
- National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China.
Organizational Affiliation: