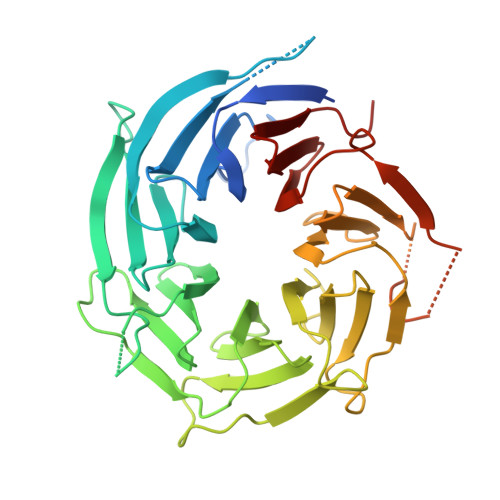
Crystal structure of human nuclear pore complex component NUP43.
Xu, C., Li, Z., He, H., Seitova, A., Wernimont, A., Li, Y., Loppnau, P., Min, J.(2015) FEBS Lett 589: 3247-3253
- PubMed: 26391640
- DOI: https://doi.org/10.1016/j.febslet.2015.09.008
- Primary Citation of Related Structures:
4I79 - PubMed Abstract:
Nuclear pore complexes (NPC) form nuclear pores that cross the nuclear envelope and allow molecules to transport between the nucleus and the cytoplasm. We solved the crystal structure of human Nup43 (hNUP43), an important component in the Nup107 subcomplex of NPC. hNup43 adopts a seven-bladed β-propeller fold. We confirmed by ITC that neither human Nup37 (hNup37) nor human Nup133 (hNup133) interacts with hNup43. We demonstrated by analytical gel filtration that the human Nup85-Seh1L binary complex recruits hNup43 to form a ternary complex. Based on amino acid sequence analysis, we predicted the hNup85-hSeh1L binding surface of hNup43.
- Structural Genomics Consortium, University of Toronto, 101 College St., Toronto, Ontario M5G 1L7, Canada; Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.
Organizational Affiliation: