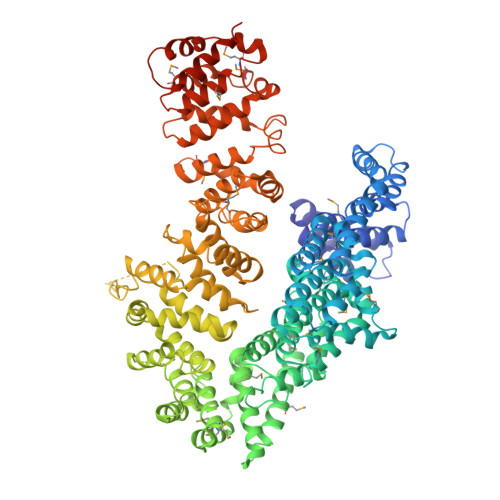
The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans.
Gazda, L., Pokrzywa, W., Hellerschmied, D., Lowe, T., Forne, I., Mueller-Planitz, F., Hoppe, T., Clausen, T.(2013) Cell 152: 183-195
- PubMed: 23332754
- DOI: https://doi.org/10.1016/j.cell.2012.12.025
- Primary Citation of Related Structures:
4I2W, 4I2Z - PubMed Abstract:
The UCS (UNC-45/CRO1/She4) chaperones play an evolutionarily conserved role in promoting myosin-dependent processes, including cytokinesis, endocytosis, RNA transport, and muscle development. To investigate the protein machinery orchestrating myosin folding and assembly, we performed a comprehensive analysis of Caenorhabditis elegans UNC-45. Our structural and biochemical data demonstrate that UNC-45 forms linear protein chains that offer multiple binding sites for cooperating chaperones and client proteins. Accordingly, Hsp70 and Hsp90, which bind to the TPR domain of UNC-45, could act in concert and with defined periodicity on captured myosin molecules. In vivo analyses reveal the elongated canyon of the UCS domain as a myosin-binding site and show that multimeric UNC-45 chains support organization of sarcomeric repeats. In fact, expression of transgenes blocking UNC-45 chain formation induces dominant-negative defects in the sarcomere structure and function of wild-type worms. Together, these findings uncover a filament assembly factor that directly couples myosin folding with myofilament formation.
- Research Institute of Molecular Pathology, Dr. Bohrgasse 7, 1030 Vienna, Austria.
Organizational Affiliation: