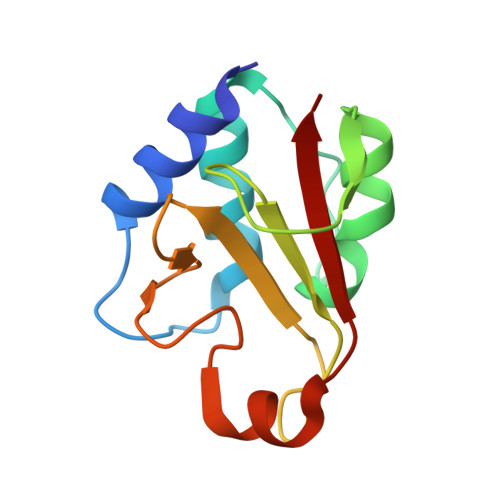
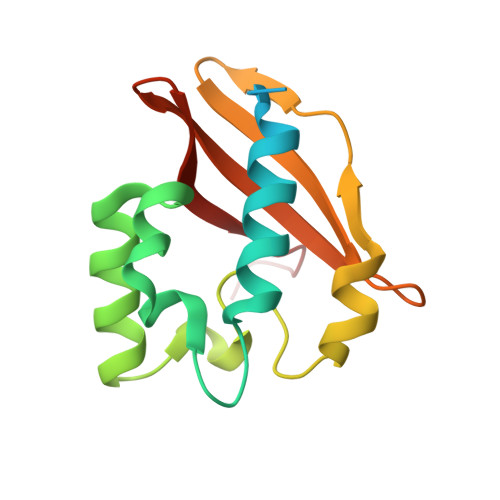
Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Dong, C., Zhang, H., Gao, Z.Q., Wang, W.J., She, Z., Liu, G.F., Shen, Y.Q., Su, X.D., Dong, Y.H.(2013) Biochem J 454: 59-68
- PubMed: 23730712
- DOI: https://doi.org/10.1042/BJ20130193
- Primary Citation of Related Structures:
4HZ9, 4HZB - PubMed Abstract:
The recently described T6SS (type VI secretion system) acts as a needle that punctures the membrane of the target cells to deliver effector proteins. Type VI amidase effectors can be classified into four divergent families (Tae1-Tae4). These effectors are secreted into the periplasmic space of neighbouring cells via the T6SS and subsequently rupture peptidoglycan. However, the donor cells are protected from damage because of the presence of their cognate immunity proteins [Tai1 (type VI amidase immunity 1)-Tai4]. In the present paper, we describe the structure of Tae3 in complex with Tai3. The Tae3-Tai3 complex exists as a stable heterohexamer, which is composed of two Tae3 molecules and two Tai3 homodimers (Tae3-Tai34-Tae3). Tae3 shares a common NlpC/P60 fold, which consists of N-terminal and C-terminal subdomains. Structural analysis indicates that two unique loops around the catalytic cleft adopt a closed conformation, resulting in a narrow and extended groove involved in the binding of the substrate. The inhibition of Tae3 is attributed to the insertion of the Ω-loop (loop of α3-α4) of Tai3 into the catalytic groove. Furthermore, a cell viability assay confirmed that a conserved motif (Gln-Asp-Xaa) in Tai3 members may play a key role in the inhibition process. Taken together, the present study has revealed a novel inhibition mechanism and provides insights into the role played by T6SS in interspecific competition.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, No. 5 Yiheyuan Road, Beijing 100871, China.
Organizational Affiliation: