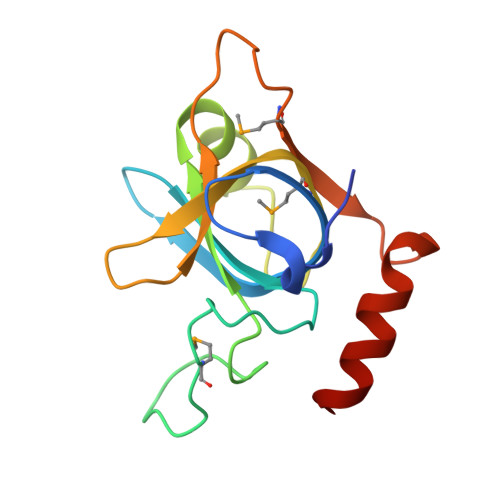
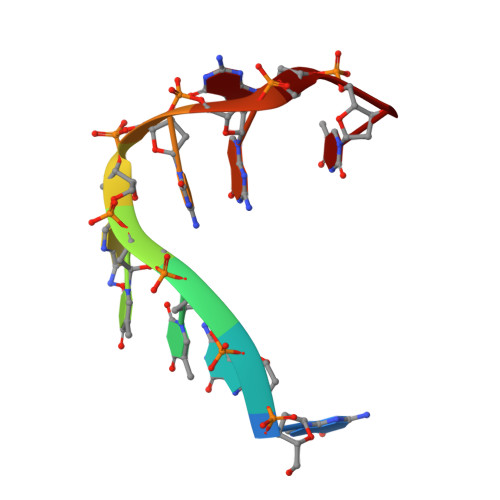Nonspecific Recognition Is Achieved in Pot1pC through the Use of Multiple Binding Modes.
Dickey, T.H., McKercher, M.A., Wuttke, D.S.(2013) Structure 21: 121-132
- PubMed: 23201273
- DOI: https://doi.org/10.1016/j.str.2012.10.015
- Primary Citation of Related Structures:
4HID, 4HIK, 4HIM, 4HIO, 4HJ5, 4HJ7, 4HJ8, 4HJ9, 4HJA - PubMed Abstract:
Pot1 is the protein responsible for binding to and protecting the 3' single-stranded DNA (ssDNA) overhang at most eukaryotic telomeres. Here, we present the crystal structure of one of the two oligonucleotide/oligosaccharide-binding folds (Pot1pC) that make up the ssDNA-binding domain in S. pombe Pot1. Comparison with the homologous human domain reveals unexpected structural divergence in the mode of ligand binding that explains the differing ligand requirements between species. Despite the presence of apparently base-specific hydrogen bonds, Pot1pC is able to bind a wide range of ssDNA sequences with thermodynamic equivalence. To address how Pot1pC binds ssDNA with little to no specificity, multiple structures of Pot1pC bound to noncognate ssDNA ligands were solved. These structures reveal that this promiscuity is implemented through new binding modes that thermodynamically compensate for base-substitutions through alternate stacking interactions and new H-bonding networks.
- Department of Chemistry and Biochemistry, University of Colorado, Boulder, CO 80309, USA.
Organizational Affiliation: