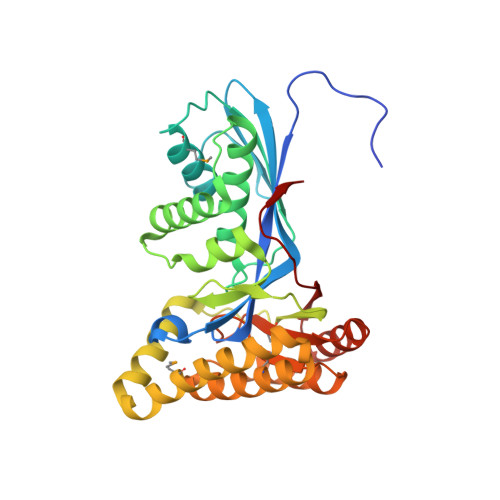
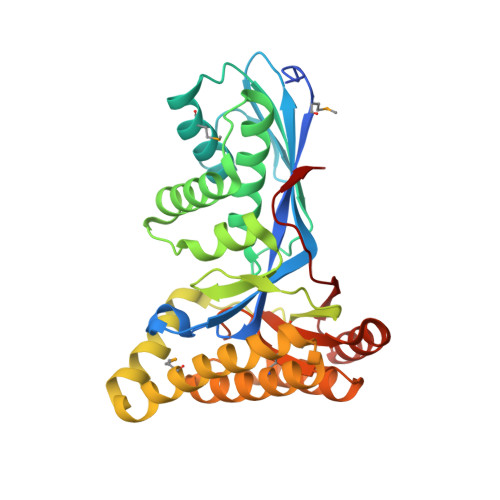
Crystallization and preliminary X-ray diffraction analysis of mevalonate kinase from Methanosarcina mazei.
Zhuang, N., Seo, K.H., Chen, C., Zhou, J., Kim, S.W., Lee, K.H.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1560-1563
- PubMed: 23192048
- DOI: https://doi.org/10.1107/S1744309112047070
- Primary Citation of Related Structures:
4HAC - PubMed Abstract:
Mevalonate kinase (MVK), which plays an important role in catalysing the biosynthesis of isoprenoid compounds derived from the mevalonate pathway, transforms mevalonate to 5-phosphomevalonate using ATP as a cofactor. Mevalonate kinase from Methanosarcina mazei (MmMVK) was expressed in Escherichia coli, purified and crystallized for structural analysis. Diffraction-quality crystals of MmMVK were obtained by the vapour-diffusion method using 0.32 M MgCl2, 0.08 M bis-tris pH 5.5, 16%(w/v) PEG 3350. The crystals belonged to space group P2(1)2(1)2, with unit-cell parameters a=97.11, b=135.92, c=46.03 Å. Diffraction data were collected to 2.08 Å resolution.
- Division of Applied Life Science (BK21 Program), Gyeongsang National University, Jinju 660-701, Republic of Korea.
Organizational Affiliation: